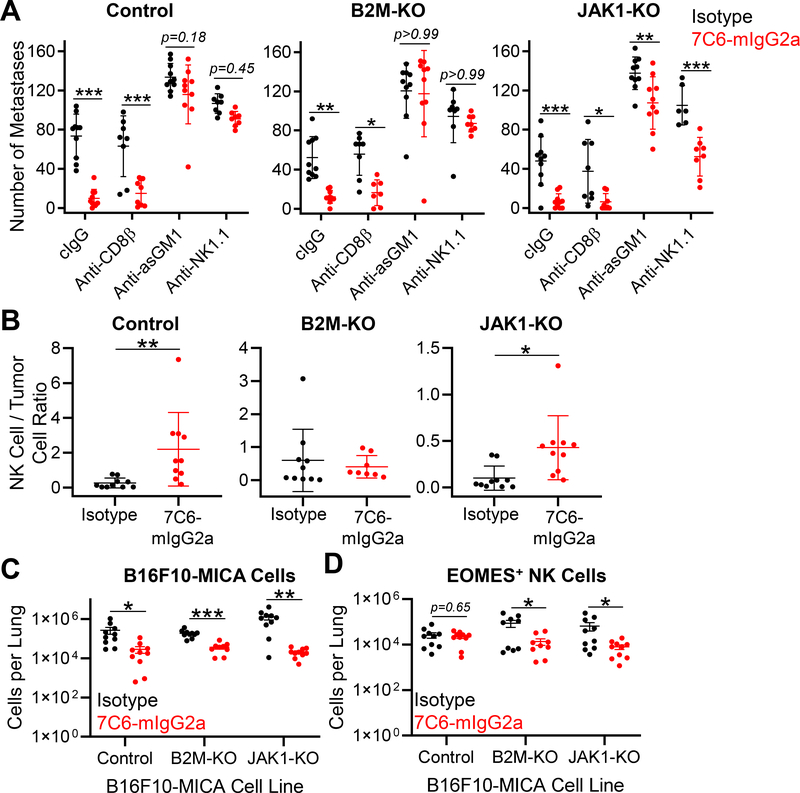
Figure 4. NK cells are essential for treatment of B2m- and Jak1-deficient melanoma metastases with a MICA/B antibody.
(A) Wild-type (WT) C57BL/6 mice were inoculated i.v. with 7×105 B16F10-MICA cells (control, B2m-KO, or Jak1-KO). Mice were treated with 7C6-mIgG2a or isotype control mAbs (200 μg) one day later, as well as on days 2 and 7. CD8+ T-cell depletion was performed by injection of anti-CD8β, whereas NK-cell depletion was performed by injection of 100μg of anti-asialo GM1 (anti-asGM1) or anti-NK1.1 on days −1, 0, and 7 after tumor cell inoculation. Control mice received an isotype control antibody. Lung surface metastases were quantified on day 14 following tumor inoculation. (B) Analysis of NK-cell infiltration into lung tissue. Tumor injection and mAb treatment were done as described in (A), with tumor cells that expressed ZsGreen to enable their identification by flow cytometry. On day 12 following tumor cell inoculation, mice were injected i.v. with APC-conjugated anti-CD45.2 to distinguish blood and tissue-infiltrating NK cells, as reported previously (23). Lung-infiltrating NK cells were identified as CD3ε–TCRβ–NK1.1+CD49b+EOMES+ viable cells with low staining for CD45.2-APC (injected i.v.) but high staining for CD45.2-PE-CY7 (added to cell suspension). The ratio of NK cells to ZsGreen+ B16F10-MICA cells is shown. (C) Numbers of ZsGreen+ B16F10-MICA cells and (D) lung-infiltrating NK cells for the indicated genotypes and treatment groups for the experiment described in (B). EOMES labeling was used to differentiate NK cells from ILC1 cells. Data pooled from two independent experiments (A-D). Statistical analyses were performed using two-way ANOVA with Bonferroni’s posthoc test (A) or two-tailed unpaired Student’s t-test (B-D), *p<0.05, **p,0.01, ***p<0.001. SD (A-B) and SEM (C-D) are shown.