Abstract
Background: Suppressor of cytokine signaling (SOCS) family proteins regulate cytokine responses through inhibition of multiple signaling pathways. The expression profiles and prognostic significance of SOCS members in ovarian cancer (OC) patients still remains unclear. Here, we aimed to provide a comprehensive understanding of the prognostic values of SOCS family members in OC and to discover promising therapeutic targets for OC. Methods: We firstly analyzed the KEGG pathway enrichment to reveal the possible pathways associated with SOCS family. Next, we used public databases including cBioPortal, Human Protein Atlas and Oncomine to investigate genetic alterations and mRNA expression of SOCS family in OC patients. More importantly, we explored the prognostic value of each individual SOCS members in OC patients using Kaplan Meier plotter database. Results: SOCS family was markedly enriched in JAK-STAT signaling pathway. A high mutation rate in SOCSs was observed in patients with ovarian serous cancer (OSC). An increased mRNA expression of SOCS1 indicated a favorable overall survival (OS) in both OC and OSC patients, while increased SOCS5 and SOCS7 expressions were significantly associated with poorer OS. We also explored the diverse roles of SOCS members in OC patients with different clinicopathological features including grades, stages and therapies employed. Conclusion: SOCS1, SOCS5 and SOCS7 may serve as prognostic biomarkers for OC patients. SOCS5 and SOCS7 may be potential therapeutic targets for OC treatment. Our results render novel insights into the association between SOCS family genes and OC development, and may highlight promising molecular targets for therapeutic interventions in OC patients.
Keywords: SOCS, ovarian cancer, prognosis, biomarker, therapeutic target
Introduction
Despite progress has been made during the past years, ovarian cancer (OC) remains the second most frequent and deadly gynecological cancer in women worldwide [1,2]. Common symptoms of OC such as bloating and pain always occur at a late stage, due to manifestation of tumor invasion and widespread intra-abdominal disease [3]; while OC is always asymptomatic at early stage. Therefore, complete surgical resection is not available for most patients at the time of diagnosis. The current main treatments for OC are cytoreductive surgery and primary chemotherapy [4]. Although chemotherapy with cisplatin plus Taxol and targeted therapy improves the survival rate of unresectable OC patients, the treatment outcomes are still disappointing. Current therapeutic options are insufficient to ensure long-term survival benefit [4,5]. Clinical remission in OC patients after treatment is commonly achievable, but over 70% of these patients will relapse, and the 5-year survival rate is less than 30% [6]. Unfortunately, OC patients respond poorly to immunotherapy which is currently seen as a type of therapeutic approach with great curative potential [7]. Radiotherapy has been largely abandoned in OC treatment [3]. To combat the huge challenges faced during the treatment of ovarian cancer, there is a growing need for identifying more specific OC biomarkers and developing novel therapeutic approaches to improve the treatment efficacy of this disease.
Inflammation is an important driving component of all cancers [8]. Proinflammatory cytokines and cytokine-activated signaling pathways are necessary contributors in inflammation and tumorigenesis [9]. As a major downstream pathway of cytokines, the Janus kinase-signal transducers and activators of transcription (JAK-STAT) signaling pathway has been aberrant activated in OC [10]. Suppressor of cytokine signaling (SOCS) family is a group of cytokine-inducible negative regulators by inhibiting multiple signaling pathways, especially the JAK/STAT signaling pathway [11-13]. This family includes eight structurally related genes, including SOCS1-SOCS7 and cytokine-inducible SH2-containing protein (CIS). Each gene consist of a central SH2 domain and a conserved C-terminus SOCS box [14]. The SH2 domain could bind to the phosphorylated tyrosine residues of cytokine receptors and JAKs, which further cause the interruption of relative signal [15]. Among all these SOCS members, SOCS1 and SOCS3 are the most potent inhibitors of JAK-STAT signaling pathway and numerous studies have demonstrated their vital roles in various malignant processes [15-17]. Considering the function of SOCS family, it is not surprising that several SOCS members may be regarded as tumor suppressor-like proteins, and dysfunction of their biology role in inhibiting cytokines and growth-factor signaling may lead to human cancer development [18].
Although the structure and function of SOCS family has been extensively studied, the relationship between expression of SOCS members and OC has not yet been clarified. Until now, there were only few reports focused on SOCS family members and OC. It is of great interest to investigate the effect of the gene expression of SOCS members on the survival of OC patients. This study is a comprehensive analysis of mRNA expression levels of SOCS family members in ovarian cancer, and with a particular focus on the prognostic values of these gene expressions. Our data revealed the potential application of SOCS family members as new biomarkers and therapeutic targets for OC prognosis and treatment. Most importantly, this is the first report about the effect of altered SOCS expression on the survival of OC patients. Furthermore, our study provides an encouraging foundation for the clinical application of novel therapeutic approaches to modulate the expression of SOCS family members in OC patients.
Materials and methods
Pathway enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) is regarded as a collection of databases that deals with a large number of genomes, biological pathways, agents, chemical materials and diseases [19]. The Database for Annotation, Visualization and Integrated Discovery (DAVID) is a commonly used online bioinformatics tool that is designed to define genes and proteins function [20]. Here, we used DAVID to visualize the SOCS family genes enrichment of pathways (P < 0.05).
cBioportal
cBioportal for Cancer Genomics (http://www.cbioportal.org/) is an open platform for interactive exploration of multidimensional Cancer Genomics data [21]. We analyzed the genetic alterations of SOCO family genes in ovarian cancer based on the Ovarian Serous Cystadenocarcinoma (TCGA, Provisional) dataset. Genomic profiles, including mutations, putative copy-number alterations, and mRNA expressions (RNA Seq V2 RSEM with z-scores = ±2) were selected for analyzing the eight SOCS family genes. The results were exhibited in the Cancer Types Summary webpage.
Human Protein Atlas
The Human Protein Atlas (HPA, http://www.proteinatlas.org) is an open access website which provides abundant data of transcriptome, proteomes and immunohistochemistry in human normal and malignant tissues [22]. We analyzed the quantitative transcriptomics data in the Pathology Atlas and obtained the fragments per kilobase of exon per million fragments mapped (FPKM) values and protein expression patterns of each SOCS gene in OC tissues.
Oncomine
Oncomine (www.oncomine.org), a cancer microarray database and web-based data mining platform, provides transcriptome data of most cancers and respective normal tissues [23]. Comparison of transcriptional expression of SOCS family genes between OC tissues and normal tissues were observed by Oncomine. mRNA level was used as data type for further analysis. p-value = 0.05 and top 10% gene rank were selected as threshold. This analysis drew on several OC studies, including TCGA Ovarian, Bonome Ovarian [24], Hendrix Ovarian [25] and Yoshihara Ovarian [26]. In addition, the 10th, 25th, 50th, 75th and 90th percentile data of each SOCS member in both cancer and normal tissues were plotted.
Kaplan-meier plotter
To evaluate the prognostic significance of the SOCS members in OC patients, we analyzed the association between gene expression and clinical outcomes through the Kaplan-Meier Plotter (www.kmplot.com). This is an online database which is capable to assess gene expression data and clinical data in specific cancer types [22,27]. Each SOCS gene was entered into the OC database and further analyzed via setting different clinical parameters. The specific Affymetrix ID of each SOCS family gene included in the Kaplan-Meier Plotter analysis were shown in Supplementary Table 1. The webpage next displayed the Kaplan Meier survival plots with hazard ratio (HR), 95% CI, and log-rank p. Patient cases were split into high and low expression groups according to the gene expression. P < 0.05 was considered to be statistically significant. We downloaded the data as text and replotted using Graphpad Prism software.
Results
Analysis of SOCS family genes via KEGG pathway enrichment
To comprehensively understand the possible pathways which the eight SOCS family genes are involved in, we initially analyzed the KEGG pathway enrichment by DAVID. The relative pathways associated with SOCS family members were summarized in Table 1. Most SOCS family members were involved in these signaling pathways. Among all these pathways, the JAK-STAT signaling pathway is the most important pathway involved in cell proliferation, survival, angiogenesis, inflammation and immune reaction [28-30]. Consistent with the results of previous investigations [18,31], almost all the SOCS genes except SOCS6 is significantly enriched in the JAK-STAT signaling pathway. Figure 1 provides a schematic illustration of the complete JAK-STAT signaling pathway and the inhibitors of this pathway including SOCS family.
Table 1.
KEGG pathway analysis of SOCS family genes
| Pathway ID | Name | Count | % | p-value | Gene |
|---|---|---|---|---|---|
| CFA04630 | JAK-STAT signaling pathway | 7 | 0.47 | 6.55e-11 | SOCS1/2/3/4/5/7, CIS |
| CFA04917 | Prolactin signaling pathway | 7 | 0.47 | 1.19e-12 | SOCS1/2/3/4/5/7, CIS |
| CAF04930 | Type II diabetes mellitus | 4 | 0.27 | 6.65e-6 | SOCS1/2/3/4 |
| CAF04910 | Insulin signaling pathway | 4 | 0.27 | 1.53e-4 | SOCS1/2/3/4 |
Figure 1.
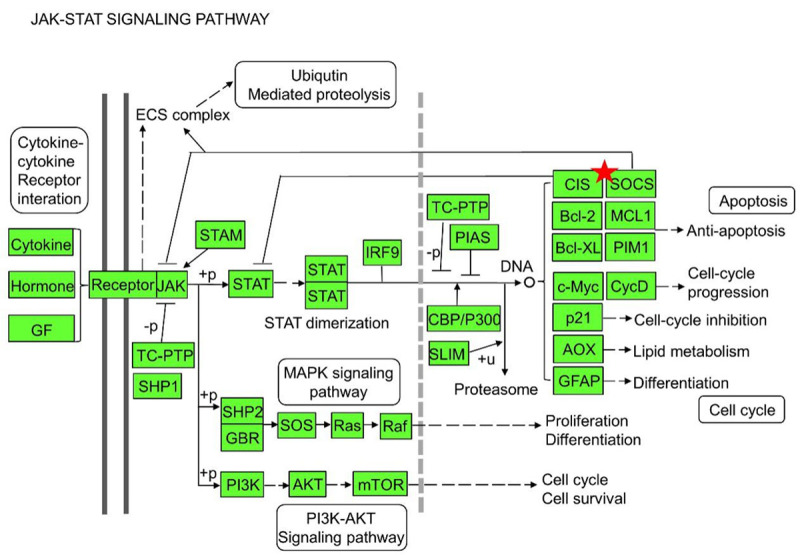
SOCS family genes were re-analyzed by KEGG pathway enrichment. The SOCS genes play an inhibiting role in the JAK-STAT signaling pathway.
Genetic mutations of SOCS family members in OC patients
Since it was unclear whether and how genetic mutations of SOCS members occur in OC, we screened the mutation type and frequency of the eight SOCS genes in serous ovarian cancer via ciBioportal. Ovarian serous cancer (OSC) is the most common histologic subtype of OC. It is classified into two major subtypes: high-grade and low-grade serous cancer, which accounts for over 70% and 10% of OC, respectively [32,33]. As shown in Figure 2A, the total mutation rate of SOCS family members observed in serous ovarian cancer patients was 48%. Aberrant expression of SOCS members included gene amplification, deep deletion, high or low mRNA expression and multiple mutations. In particular, mRNA upregulation was the most common alteration.
Figure 2.

Genetic alterations and gene expression of SOCS family genes in OC. A. Alteration frequency of SOCSs in OC via cBioPortal. B. The FPKM values of SOCS family members in OC via HPA.
Next, we analyzed the mRNA expression of individual SOCS members in OC via Pathology Atlas of HPA database. The SOCS3 and SOCS1 genes exhibited relative high level of mRNA expression among all the genes, on the contrary, SOCS2 mRNA expression level was the lowest (Figure 2B). In addition, we also investigated the immunohistochemistry data of HPA database to assess the protein level of each SOCS members. As summarized in Supplementary Table 2, most SOCS genes except SOCS6 were relatively highly expressed in OC tissues. However, the data was not strongly convincing to explain the protein expression of SOCSs in OC due to the insufficient sample size.
mRNA expression of SOCS family members in OC patients
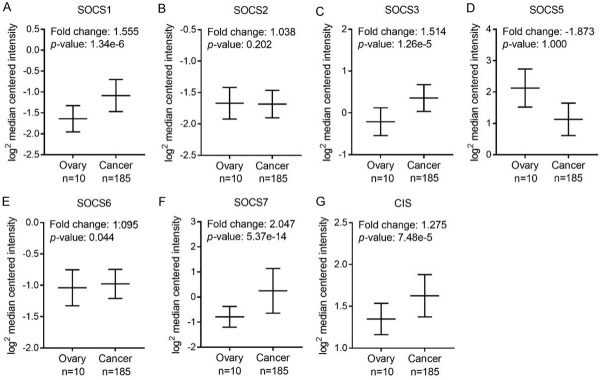
To compare the mRNA expression of SOCS members in ovarian cancer and normal tissues, we explored the OC and OSC data respectively from Bonome ovarian dataset [24] and TCGA dataset. As shown in Figure 3, almost all the SOCS members except SOCS5 is upregulated in OC tissues. Specifically, the increased expression of SOCS1, SOCS3, SOCS6, SOCS7 and CIS in OC tissues compared with that of the normal controls showed statistical significance. We next analyzed the mRNA level of SOCSs in OSC compared with that of the normal controls. The mRNA expression of SOCS1, SOCS3, SOCS5 and SOCS7 was significantly higher in OSC tissues (Figure 4A, 4C, 4D and 4F), whereas SOCS2, SOCS6 and CIS are either slightly upregulated or downregulated without a statistically significant p value (Figure 4B, 4E and 4G). No available data about SOCS4 expression level between OC or OSC tissues and normal tissues.
Figure 3.
The mRNA expression of each SOCS member in ovarian tissues in Bonome dataset. Comparasion of SOCS1 (A), SOCS2 (B), SOCS3 (C), SOCS5 (D), SOCS6 (E), SOCS7 (F), CIS (G) mRNA expression in normal ovarian tissue (left plot, n = 10) and ovarian cancer tissue (right plot, n = 185) using Oncomine.
Figure 4.
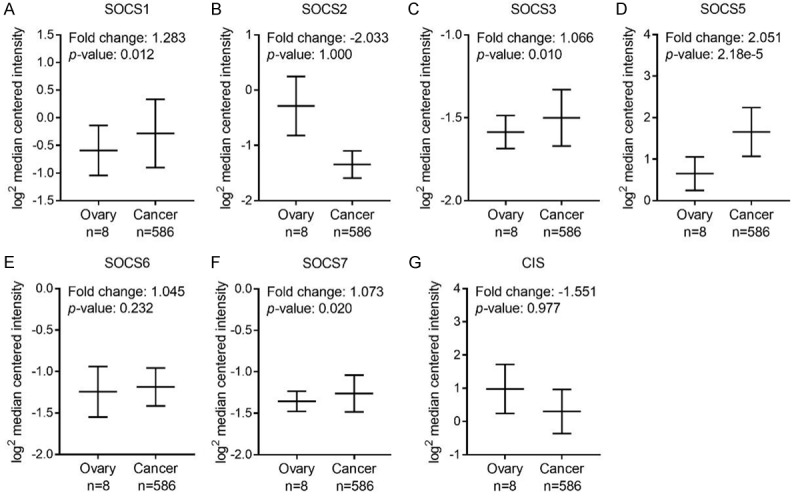
The mRNA expression of each SOCS member in ovarian tissues in TCGA dataset. Comparasion of SOCS1 (A), SOCS2 (B), SOCS3 (C), SOCS5 (D), SOCS6 (E), SOCS7 (F), CIS (G) mRNA expression in normal ovarian tissue (left plot, n = 8) and ovarian cancer tissue (right plot, n = 586) using Oncomine.
In addition, we explored mRNA expression of each SOCS members in Hendrix Ovarian [25] and Yoshihara Ovarian [26] datasets. No expression of significant difference was observed in these two datasets (Table 2). The inconsistent results obtained from these datasets probably attribute to its different sample size, study design as well as detection methods.
Table 2.
The mRNA expression of SOCS family genes in ovarian serous cancer compared with normal control
| Gene | p-value | Fold Chang | Dataset | #Normal | #Cancer |
|---|---|---|---|---|---|
| SOCS1 | 0.939 | -1.096 | Hendrix Ovarian | 4 | 41 |
| 1.000 | -1.778 | Yoshihara Ovarian | 10 | 43 | |
| SOCS2 | 1.000 | -1.437 | Hendrix Ovarian | 4 | 41 |
| 1.000 | -49.946 | Yoshihara Ovarian | 10 | 43 | |
| SOCS3 | 0.082 | 1.036 | Hendrix Ovarian | 4 | 41 |
| 1.000 | -13.902 | Yoshihara Ovarian | 10 | 43 | |
| SOCS4 | 0.335 | 1.081 | Yoshihara Ovarian | 10 | 43 |
| SOCS5 | 0.998 | -1.152 | Hendrix Ovarian | 4 | 41 |
| 1.000 | -2.036 | Yoshihara Ovarian | 10 | 43 | |
| SOCS6 | 0.105 | 1.018 | Hendrix Ovarian | 4 | 41 |
| 0.900 | -1.524 | Yoshihara Ovarian | 10 | 43 | |
| SOCS7 | 0.150 | 1.080 | Hendrix Ovarian | 4 | 41 |
| 0.760 | -1.027 | Hendrix Ovarian | 4 | 41 | |
| CIS | 1.000 | -1.923 | Yoshihara Ovarian | 10 | 43 |
Prognostic values of SOCS family members in OC patients
To reveal the prognostic values of SOCS family members in OC patients, we analyzed the Kaplan-Meier Plotter database and plotted the OS curves. Among the eight SOCS genes, only an increased mRNA expression level of SOCS1 was significantly associated with favorable OS (Figure 5A, HR = 0.83, 95% CI: 0.72-0.95, P = 0.0079). Increased mRNA expression of SOCS2, SOCS5 and SOCS7 indicated poor OS (Figure 5B, 5E and 5G; SOCS2, HR = 1.32, 95% CI: 1.08-1.62, P = 0.0064; SOCS5, HR = 1.46, 95% CI: 1.28-1.66, P = 1.2e-08; SOCS7, HR = 1.41 95% CI: 1.15-1.73, P = 0.001). The mRNA expression of other SOCS family members had no association with the OS of OC patients (Figure 5C, 5D, 5F and 5H).
Figure 5.
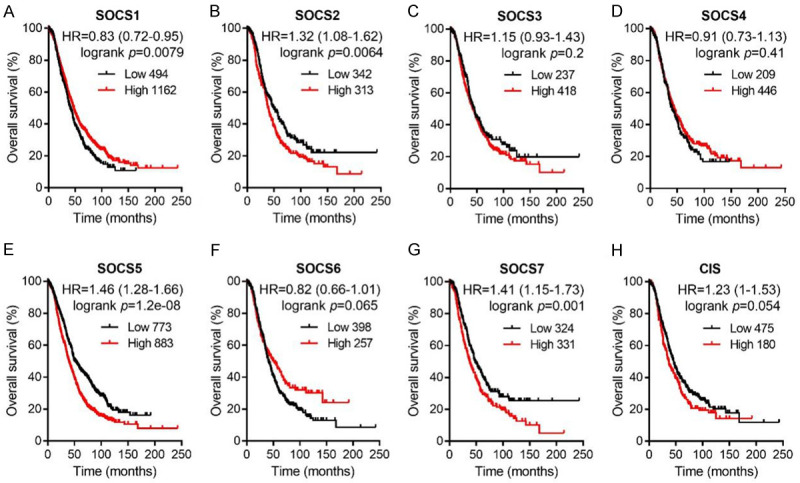
Survival analysis of SOCS1 (A, n = 1656), SOCS2 (B, n = 655), SOCS3 (C, n = 655), SOCS4 (D, n = 655), SOCS5 (E, n = 1656), SOCS6 (F, n = 655), SOCS7 (G, n = 655), CIS (H, n = 655) in all OC patients. Data was analyzed by Kaplan-Meier plotter. Patients with expression above the median are indicated in red line, and patients with expressions below the median are indicated in black line. HR means hazard ratio.
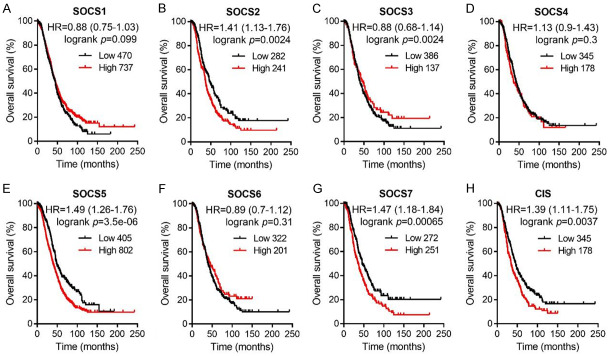
We also evaluated the prognostic values of SOCS mRNA expression in OSC patients. As shown in Figure 6, a higher mRNA expressions level of SOCS2, SOCS5, SOCS7 and CIS was significantly related to a shorter OS for OSC patients (SOCS2, HR = 1.41, 95% CI: 1.13-1.76, P = 0.0024; SOCS5, HR = 1.49, 95% CI: 1.26-1.76, P = 3.5e-06; SOCS7, HR = 1.47, 95% CI: 1.18-1.84, P = 0.00065; CIS, HR = 1.39, 95% CI: 1.11-1.75, P = 0.0037). There was no significant correlation between the residual SOCS family members and the OS of OSC patients. Considered that SOCS2 mRNA expression was low in ovarian cancer tissues and the FPKM value of SOCS2 in OC was the lowest, we would not regard SOCS2 as a prognostic biomarker in OC patients. Therefore, it is reasonable to suggest that SOCS5 and SOCS7 targeted agents may be attractive candidates for the treatment of OC. Furthermore, we analyzed the effect of SOCS family members on the progression-free survival (PFS) rates in OC as well as OSC patients (Supplementary Figures 1 and 2). Taken together, these data provided a basis for further studies of the mechanism of different SOCS family members in the inhibition and development of OC.
Figure 6.
Survival analysis of SOCS1 (A, n = 1207), SOCS2 (B, n = 523), SOCS3 (C), SOCS4 (D, n = 523), SOCS5 (E, n = 1207), SOCS6 (F, n = 523), SOCS7 (G, n = 523), CIS (H, n = 523) in OSC patients. Data was analyzed by Kaplan-Meier plotter. Patients with expression above the median are indicated in red line, and patients with expressions below the median are indicated in black line. HR means hazard ratio.
Prognostic values of SOCSs in OC patients with different clinicopathological features
To confirm whether the prognostic effect of SOCS mRNA expression on the survival of OC patients was associated with different clinicopathological features, we evaluated the prognostic values of SOCS members in OC patients with different TP53 status, clinical stages, pathological grades and therapies employed via the Kaplan-Meier plotter database. Firstly, we found that an overexpression of SOCS3 and CIS indicated a positive OS in TP53 mutated and wide type OC patients, respectively (Table 3). SOCS5 and SOCS6 overexpression was associated with negative OS in both TP53 wide type and mutated OC patients. Increased mRNA expression level of SOCS2 was related to negative OS in TP53 mutated OC patients.
Table 3.
Correlation of SOCS family members with TP53 status in OC patients
| Gene | TP53 status | Case-low1 | Case-high2 | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| SOCS1 | wide type | 29 | 65 | 1.63 (0.9-2.94) | 0.1 |
| mutated | 377 | 129 | 1.29 (1-1.66) | 0.049* | |
| SOCS2 | wide type | 8 | 11 | 2.87 (0.9-9.12) | 0.063 |
| mutated | 71 | 53 | 1.48 (1.01-2.17) | 0.043* | |
| SOCS3 | wide type | 9 | 10 | 0.69 (0.25-1.92) | 0.48 |
| mutated | 42 | 82 | 0.66 (0.45-0.98) | 0.039* | |
| SOCS4 | wide type | 7 | 12 | 2.28 (0.76-6.82) | 0.13 |
| mutated | 36 | 88 | 0.68 (0.45-1.02) | 0.058 | |
| SOCS5 | wide type | 23 | 71 | 2.58 (1.2-5.53) | 0.012* |
| mutated | 126 | 380 | 1.23 (0.94-1.62) | 0.12 | |
| SOCS6 | wide type | 8 | 11 | 3.15 (1.05-9.46) | 0.032* |
| mutated | 90 | 34 | 1.34 (0.88-2.04) | 0.18 | |
| SOCS7 | wide type | 5 | 14 | 1.57 (0.49-4.96) | 0.44 |
| mutated | 49 | 75 | 0.68 (0.46-0.99) | 0.045* | |
| CIS | wide type | 7 | 12 | 0.26 (0.08-0.82) | 0.014* |
| mutated | 34 | 90 | 1.47 (0.95-2.28) | 0.082 |
Cases-low: patient number-low expression of the corresponding gene.
Cases-high: patient number-high expression of the corresponding gene.
P < 0.05.
Detailed analysis of SOCS expression with its respective OC stages and grades were shown in Tables 4 and 5. The data in Table 4 revealed that SOCS6 and CIS was associated with better OS in Stage I and Stage II OC patients, while SOCS2 indicated poor survival. SOCS1 expression level was related to favorable OS in Stage III and Stage IV OC patients. Meanwhile, SOCS5, SOCS6, SOCS7 and CIS were correlated with unfavorable OS in these patients. In Table 5, we exhibited the prognostic value between mRNA expression of SOCS members and different histological grades. Notably, an increased mRNA expression of SOCS5 was associated with poor OS in all the OC patients. SOCS3 indicated poor OS in Grade I and Grade II OC patients while SOCS2 indicated poor OS in Grade III OC patients.
Table 4.
Correlation of mRNA expression of SOCS family members with tumor stages of OC patients
| Gene | Stages | Case-low1 | Case-high2 | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| SOCS1 | 1 + 2 | 91 | 44 | 0.53 (0.2-1.4) | 0.19 |
| 3 + 4 | 585 | 635 | 0.82 (0.71-0.95) | 0.0094** | |
| SOCS2 | 1 + 2 | 30 | 53 | 4.26 (0-inf) | 0.0015** |
| 3 + 4 | 154 | 333 | 0.87 (0.68-1.1) | 0.25 | |
| SOCS3 | 1 + 2 | 54 | 29 | 0.28 (0.06-1.25) | 0.074 |
| 3 + 4 | 363 | 124 | 0.81 (0.63-1.06) | 0.13 | |
| SOCS4 | 1 + 2 | 61 | 22 | 1.76 (0.62-4.98) | 0.28 |
| 3 + 4 | 331 | 156 | 1.13 (0.89-1.44) | 0.33 | |
| SOCS5 | 1 + 2 | 68 | 67 | 1.57 (0.71-3.47) | 0.26 |
| 3 + 4 | 460 | 760 | 1.49 (1.27-1.75) | 5.4e-07** | |
| SOCS6 | 1 + 2 | 21 | 62 | 0.23 (0.08-0.64) | 0.0022** |
| 3 + 4 | 216 | 271 | 1.33 (1.06-1.67) | 0.015* | |
| SOCS7 | 1 + 2 | 30 | 53 | 0.46 (0.17-1.28) | 0.13 |
| 3 + 4 | 257 | 230 | 1.26 (1.01-1.58) | 0.045* | |
| CIS | 1 + 2 | 34 | 49 | 0.35 (0.12-1) | 0.041* |
| 3 + 4 | 354 | 133 | 1.28 (1.01-1.63) | 0.041* |
Cases-low: patient number-low expression of the corresponding gene.
Cases-high: patient number-high expression of the corresponding gene.
P < 0.05;
P < 0.01.
Table 5.
Correlation of SOCS family members with tumor grades of OC patients
| Gene | Grades | Case-low1 | Case-high2 | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| SOCS1 | I + II | 206 | 174 | 0.62 (0.46-0.83) | 0.0011** |
| III | 451 | 169 | 0.51 (0.35-0.76) | 0.00064** | |
| IV | 12 | 8 | 0.58 (0.21-1.55) | 0.27 | |
| SOCS2 | I + II | 139 | 64 | 0.77 (0.5-1.18) | 0.23 |
| III | 136 | 68 | 1.28 (0.76-2.16) | 0.34 | |
| IV | 3 | 15 | 1219678945.7(0-lnf) | 0.0043* | |
| SOCS3 | I + II | 81 | 122 | 1.72 (1.1-2.68) | 0.016* |
| III | 128 | 76 | 0.66 (0.38-1.14) | 0.13 | |
| IV | 10 | 8 | 0.42 (0.14-1.26) | 0.11 | |
| SOCS4 | I + II | 110 | 93 | 0.78 (0.52-1.18) | 0.24 |
| III | 53 | 151 | 0.48 (0.28-0.82) | 0.0055* | |
| IV | 4 | 14 | 2.5 (0.68-9.21) | 0.16 | |
| SOCS5 | I + II | 202 | 178 | 1.79 (1.34-2.4) | 6.3e-05** |
| III | 389 | 231 | 1.52 (1.13-2.03) | 0.0047* | |
| IV | 14 | 6 | 3.45 (1.08-10.97) | 0.026* | |
| SOCS6 | I + II | 131 | 72 | 0.61 (0.38-0.96) | 0.032* |
| III | 105 | 99 | 0.58 (0.34-0.98) | 0.038* | |
| IV | 13 | 5 | 0.53 (0.15-1.88) | 0.32 | |
| SOCS7 | I + II | 62 | 141 | 1.45 (0.91-2.33) | 0.12 |
| III | 147 | 57 | 1.59 (0.95-2.67) | 0.076 | |
| IV | 12 | 6 | 1.87 (0.65-5.38) | 0.24 | |
| CIS | I + II | 118 | 85 | 0.65 (0.43-0.99) | 0.043* |
| III | 91 | 71 | 0.7 (0.45-1.1) | 0.12 | |
| IV | 11 | 7 | 0.4 (0.13-1.28) | 0.11 |
Cases-low: patient number-low expression of the corresponding gene.
Cases-high: patient number-high expression of the corresponding gene.
P < 0.05;
P < 0.01.
The results in Tables 6 and 7 showed the correlation between mRNA level of SOCSs and OS in OC patients who underwent chemotherapy or surgery. As shown in Table 6, high expression of SOCS2, SOCS5, SOCS7 and CIS predicted poor OS in patients receiving platin-based chemotherapy. An increased level of SOCS7 expression indicated an unfavorable OS in patients receiving taxol-based chemotherapy. Moreover, higher expression of SOCS3 and SOCS5 was related to shorter OS in OC patients with complete surgical resection, while an increased expression of SOCS6 and CIS indicated better OS (Table 7). The data that focused on OC patients who received cytoreductive surgery has demonstrated that high expression of SOCS2, SOCS6 and SOCS7 was correlated with unfavorable OS.
Table 6.
Correlation of SOCS members with OC patients receiving different chemotherapy regimes
| Gene | Chemotherapy | Case-low1 | Case-high2 | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| SOCS1 | contains platin | 505 | 904 | 0.84 (0.73-0.97) | 0.017* |
| contains taxol | 318 | 475 | 0.71 (0.59-0.86) | 0.00037** | |
| SOCS2 | contains platin | 262 | 216 | 1.3 (1.03-1.64) | 0.026* |
| contains taxol | 198 | 159 | 1.24 (0.94-1.66) | 0.13 | |
| SOCS3 | contains platin | 287 | 191 | 1.24 (0.98-1.57) | 0.07 |
| contains taxol | 221 | 136 | 1.22 (0.91-1.63) | 0.19 | |
| SOCS4 | contains platin | 260 | 218 | 0.89 (0.71-1.13) | 0.33 |
| contains taxol | 198 | 159 | 0.82 (0.61-1.09) | 0.17 | |
| SOCS5 | contains platin | 679 | 730 | 1.41 (1.22-1.62) | 1.8e-06** |
| contains taxol | 416 | 377 | 1.48 (1.23-1.79) | 3.5e-05** | |
| SOCS6 | contains platin | 125 | 353 | 1.15 (0.88-1.5) | 0.3 |
| contains taxol | 239 | 118 | 0.79 (0.57-1.09) | 0.15 | |
| SOCS7 | contains platin | 269 | 209 | 1.33 (1.05-1.67) | 0.016* |
| contains taxol | 239 | 118 | 1.38 (1.03-1.85) | 0.03* | |
| CIS | contains platin | 353 | 125 | 1.32 (1.03-1.69) | 0.03* |
| contains taxol | 267 | 90 | 1.31 (0.96-1.79) | 0.091 |
Cases-low: patient number-low expression of the corresponding gene.
Cases-high: patient number-high expression of the corresponding gene.
P < 0.05;
P < 0.01.
Table 7.
Correlation of SOCS family members with debulk status of OC patients
| Gene | Debulk | Case-low1 | Case-high2 | HR (95% CI) | p-value |
|---|---|---|---|---|---|
| SOCS1 | optimal | 245 | 556 | 0.85 (0.68-1.05) | 0.13 |
| suboptimal | 213 | 323 | 0.8 (0.65-0.98) | 0.03* | |
| SOCS2 | optimal | 60 | 183 | 1.34 (0.81-2.21) | 0.26 |
| suboptimal | 117 | 118 | 1.52 (1.13-2.05) | 0.0055** | |
| SOCS3 | optimal | 112 | 131 | 1.73 (1.13-2.64) | 0.01* |
| suboptimal | 59 | 176 | 0.75 (0.54-1.05) | 0.096 | |
| SOCS4 | optimal | 166 | 77 | 1.45 (0.95-2.21) | 0.082 |
| suboptimal | 161 | 74 | 0.76 (0.55-1.06) | 0.11 | |
| SOCS5 | optimal | 383 | 418 | 1.7 (1.38-2.09) | 3.7e-07** |
| suboptimal | 307 | 229 | 1.28 (1.05-1.57) | 0.015* | |
| SOCS6 | optimal | 172 | 71 | 0.4 (0.23-0.7) | 0.001** |
| suboptimal | 174 | 61 | 1.41 (1.01-1.95) | 0.041* | |
| SOCS7 | optimal | 170 | 73 | 1.17 (0.76-1.8) | 0.47 |
| suboptimal | 170 | 65 | 1.58 (1.15-2.18) | 0.0045** | |
| CIS | optimal | 158 | 85 | 0.62 (0.4-0.96) | 0.031* |
| suboptimal | 116 | 119 | 1.38 (1.02-1.86) | 0.0371* |
Cases-low: patient number-low expression of the corresponding gene.
Cases-high: patient number-high expression of the corresponding gene.
P < 0.05;
P < 0.01.
Discussion
OC remains a big challenge to clinicians and researches due to its clinical outcomes and poor prognosis [34]. Despite increasing attention on OC treatment, a better understanding of the molecular biology of this tumor and how to improve therapy remains required urgently. In addition, oncological treatment needs to be individualized to ensure optimal efficiency of therapy. SOCS is a big family of proteins that are commonly regarded as antagonists of JAK-STAT signaling and regulators of other signaling pathways. There is indeed a positive correlation between SOCS dysregulation and tumor progression [18]. However, studies about SOCS and OC are few in number and the specific function of SOCS proteins in OC has not been determined. Here, we integrated several publicly available data into one major analysis to explore the prognostic value of SOCS members in OC for the first time.
SOCS1 was found to participate in the regulation of signaling pathways such as NF-κB, STAT FAK, p38 MAPK and p53 pathways [35-37]. SOCS1 gene silencing was observed in human hepatic carcinoma, gastric cancer, multiple melanoma and pancreatic cancer [38-41]. On the other hand, SOCS1 overexpression exhibited a potent antitumor effect in malignant pleural mesothelioma [42], and also sensitized glioblastoma as well as cervical cancer cells to radiation therapy [43,44]. These suggested that SOCS1 activator might be a promising agent for cancer treatment. The role of SOCS1 in OC has not yet been explored. Our data revealed that the mRNA level of SOCS1 was up-regulated in OC tissues compared with that of normal controls and low SOCS1 mRNA expression level was significantly associated with unfavorable prognosis in OC patients. This hinted that overexpression of SOCS1 was also required for growth inhibition of OC. Thus, we speculated that inhibition of JAK-STAT3 and other signaling pathways might be attributable to the molecular mechanisms underlying the inhibition effect of SOCS1 on OC cell growth. Another possible mechanism reported in colorectal cancer was that SOCS1 prevented epithelial-mesenchymal transition and promoted mesenchymal-epithelial transition to further inhibit invasion and migration of cancer cells. Mechanistic research needs to illustrate the underlying process by which SOCS1 inhibits OC cell survival.
The expression of SOCS2 in hepatocellular carcinoma was significantly lower than that in normal liver tissues and patients with low SOCS2 expression had significantly shorter OS [45]. Breast cancer patients with high SOCS2 expression lived significantly longer lives [46]. Decreased SOCS2 expression in prostate cancer was associated with an increased incidence of metastasis [47]. Reduced SOCS2 expression always happened during the adenoma to colorectal cancer sequence [48]. In contrast, another research provided the evidence that SOCS2 exerted growth promoting effects in prostate cancer [49]. This conclusion was consistent with our data. One investigation that focused on SOCS2 and OC only reported that hypermethylation of SOCS2 has been detected in OC [50]. These controversial conclusions might be due to different tumor types. Our data revealed that high mRNA expression of SOCS2 was associated with poor survival of OC patients. However, the mRNA level and FPKM value of SOCS2 was lower in ovarian malignant tissues in both On-comine and HPA database analysis. We did not consider SOCS2 as a prognostic indicator in our study. It is worth noting that the protein expression of SOCS2 in OC tissue was maintained at high and medium levels. However, the sample size of these immunohistochemical data from HPA was inadequate.
SOCS3 is structurally similar with SOCS1 and both of them are JAK kinase strong inhibitors due to the unique KIR domain, which is critical for JAK inhibition [51]. SOCS3 participates in tumor development and progression, but its functions are highly dependent on tumor types. SOCS3 was undetectable in human lung, liver, and head and neck cancers, which provided a growth advantage for tumor cells [52-54]. In contrast, SOCS3 that was expressed in breast cancer, prostate cancer and glioblastoma, further promoted cancer cell growth and even enhanced the resistance of glioblastoma to radiation therapy [43,55,56]. Reduced expression or silencing of SOCS3 was considered having a correlation with constitutive STAT3 activation, which in turn promoted tumorigenesis via activating tumor-promoting genes [57]. In our study, mRNA expression of SOCS3 exhibited no significant connection to the survival of OC patients.
Several researches have already found that SOCS4 could suppress tumor growth [58,59]. High mRNA expression of SOCS4 was significantly associated with better clinical outcome in breast cancer [60]. In contrast, SOCS4 hypermethylation indicated a poor prognosis for GC patients [58]. Although SOCS4 expression is significantly higher in ovarian cancer tissues than that in normal controls, there is no correlation between SOCS4 expression and overall survival in OC patients. Interestingly, OC patients with increased SOCS4 expression had a better progression-free survival.
It was generally accepted that SOCS5 possessed the tumor suppression ability [44,61]. SOCS5 inhibited the invasion and metastasis of pancreatic cancer by inactivating the JAK/STAT3 signaling pathway [60,62]. SOCS5 negatively regulated cell growth and cell cycle progression in T-cell lineage acute lymphoblastic leukemia [63]. In addition, SOCS5 reacted against the maintenance of cancer stem cell characteristics and chemoresistance in hepatocellular carcinoma [64]. On the contrary, recent study found that SOCS5 was significantly overexpressed in hepatocellular carcinoma tissues, and its overexpression caused tumor cell migration and invasion by blocking PI3K/Akt/mTOR-mediated autophagy [65]. There are more studies that revealed the conflicting tumor-promoting and tumor-suppressive roles of SOCS5. The biological function of SOCS5 in multiple cancers is critical yet incompletely understood. Our data showed that SOCS5 might act as a promotor and strongly support ovarian tumor progression. It suggested that SOCS5 might be used as therapy target for OC treatment. Further studies are needed to shed more light on the growth-promoting mechanisms of SOCS5 in OC.
SOCS6, known as an important regulator in insulin signaling, was reduced in many cancers such as colorectal cancer, prostate cancer, hepatocellular cancer [45,48,66]. SOCS6 was frequently down-regulated and showed a suppressive role in gastric cancer [67]. SOCS6 also participated in suppression of cell growth and metastasis in Kaposi’s sarcoma cells [68]. In our study, a significant correlation was found between SOCS6 expression and survival of OC patients. However, we found that the prognostic values of SOCS6 in early stage and advanced stage patients are inconsistent. SOCS6 overexpression was associated with a favorable OS in early stage patients, while its overexpression indicated poor OS in advanced stage patients. These data demonstrated that SOCS6 might be involved in ovarian tumor progression.
Few investigations are currently available about the functions of SOCS7 in multiple human tumors. Increased SOCS7 mRNA expression was related to a favorable prognosis in human breast cancer [60]. An increased expression of SOCS7 impaired the aggressive action of prostate cancer cells through terminating the JAK-STAT3 pathway activation [69]. Here, we found that SOCS7 was highly expressed in OC patients compared with that of the normal control, and increased level of expression of this gene was significantly associated with unfavorable survival of OC patients. Our results laid a foundation for the potential possibility of SOCS7 regarded as a promising therapeutic target for OC treatment.
In addition, we also investigated the relationship between SOCS members and OC patients with different therapeutic strategies. Specific SOCS family members could be used to predict the prognosis of OC patients. Hence, different chemotherapy regimens and different types of surgeries could be designed for the patients. According to the results of our data, it is likely that inhibiting the expression of some SOCS member may provide a possibility to improve the survival rate of OC patients. Therefore, chemotherapy or surgery combine with specific SOCS targeted agents such as a SOCS activator or inhibitor might improve therapeutic efficacy for OC. More attention should be given to achieve this promising and meaningful clinical possibility.
The current treatment outcomes of OC remain disappointing, it is highly necessary to find clinical and molecular determinants associated with the outcome of OC to develop novel therapeutic approaches. Our bioinformatics study explained the prognostic values of SOCS family members in ovarian cancer. Specifically, SOCS1, SOCS5 and SOCS7 displayed an increased expression at the mRNA level in OC patients and overexpression of these genes is significantly associated with the survival of OC patients. Therefore, SOCS1 could potentially be an effective prognostic biomarker for improving the survival of OC patients. On another hand, targeting SOCS5 and SOCS7 may provide a potential therapeutic approach for OC treatment. Furthermore, overexpression of SOCS1 might exhibit a potent antitumor effect against OC. Although the protein level of SOCSs and the detailed mechanisms need further investigations to clarify, our present data validated and supported bioinformatics as an appropriate starting point for analyzing human ovarian cancer and discovering novel biomarkers and therapeutic targets for OC.
Acknowledgements
We are very grateful to the contributors of data to KEGG, cBioPortal, HPA, Oncomine, and Kaplan-Meier plotter. This study was supported by the National Natural Science Foundation of China (No. 81501137; 81971805; 81672619) and Guangzhou Science and Technology Research Project (No. 201804010061).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Herrera FG, Irving M, Kandalaft LE, Coukos G. Rational combinations of immunotherapy with radiotherapy in ovarian cancer. Lancet Oncol. 2019;20:e417–e433. doi: 10.1016/S1470-2045(19)30401-2. [DOI] [PubMed] [Google Scholar]
- 4.Kyrgiou M, Salanti G, Pavlidis N, Paraskevaidis E, Ioannidis JP. Survival benefits with diverse chemotherapy regimens for ovarian cancer: meta-analysis of multiple treatments. J Natl Cancer Inst. 2006;98:1655–1663. doi: 10.1093/jnci/djj443. [DOI] [PubMed] [Google Scholar]
- 5.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, Sonke GS, Gourley C, Banerjee S, Oza A, Gonzalez-Martin A, Aghajanian C, Bradley W, Mathews C, Liu J, Lowe ES, Bloomfield R, DiSilvestro P. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 6.Matz M, Coleman MP, Carreira H, Salmeron D, Chirlaque MD, Allemani C CONCORD Working Group. Erratum to “Worldwide comparison of ovarian cancer survival: histological group and stage at diagnosis (CONCORD-2)” [Gynecol. Oncol. 144 (2017) 396-404] . Gynecol Oncol. 2017;147:725. doi: 10.1016/j.ygyno.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coukos G, Tanyi J, Kandalaft LE. Opportunities in immunotherapy of ovarian cancer. Ann Oncol. 2016;27(Suppl 1):i11–i15. doi: 10.1093/annonc/mdw084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 9.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burke WM, Jin X, Lin HJ, Huang M, Liu R, Reynolds RK, Lin J. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20:7925–7934. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 11.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 12.Cooney RN. Suppressors of cytokine signaling (SOCS): inhibitors of the JAK/STAT pathway. Shock. 2002;17:83–90. doi: 10.1097/00024382-200202000-00001. [DOI] [PubMed] [Google Scholar]
- 13.McCormick SM, Heller NM. Regulation of macrophage, dendritic cell, and microglial phenotype and function by the SOCS proteins. Front Immunol. 2015;6:549. doi: 10.3389/fimmu.2015.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piessevaux J, Lavens D, Peelman F, Tavernier J. The many faces of the SOCS box. Cytokine Growth Factor Rev. 2008;19:371–381. doi: 10.1016/j.cytogfr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki-Ohara K, Kondo T, Ito M, Yoshimura A. SOCS, inflammation, and cancer. JAKSTAT. 2013;2:e24053. doi: 10.4161/jkst.24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inagaki-Ohara K, Mayuzumi H, Kato S, Minokoshi Y, Otsubo T, Kawamura YI, Dohi T, Matsuzaki G, Yoshimura A. Enhancement of leptin receptor signaling by SOCS3 deficiency induces development of gastric tumors in mice. Oncogene. 2014;33:74–84. doi: 10.1038/onc.2012.540. [DOI] [PubMed] [Google Scholar]
- 17.Kamio M, Yoshida T, Ogata H, Douchi T, Nagata Y, Inoue M, Hasegawa M, Yonemitsu Y, Yoshimura A. SOCS1 [corrected] inhibits HPV-E7-mediated transformation by inducing degradation of E7 protein. Oncogene. 2004;23:3107–3115. doi: 10.1038/sj.onc.1207453. [DOI] [PubMed] [Google Scholar]
- 18.Wu M, Song D, Li H, Yang Y, Ma X, Deng S, Ren C, Shu X. Negative regulators of STAT3 signaling pathway in cancers. Cancer Manag Res. 2019;11:4957–4969. doi: 10.2147/CMAR.S206175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M, Chen H, Zhou L, Chen K, Su F. Expression profile and prognostic values of STAT family members in non-small cell lung cancer. Am J Transl Res. 2019;11:4866–4880. [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC, Boyd J, Birrer MJ. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 26.Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, Kudo Y, Inoue I, Tanaka K. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng WX, Huang JG, Yang L, Gong AH, Mo YY. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol Cancer. 2017;16:161. doi: 10.1186/s12943-017-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. 2013;12:611–629. doi: 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pencik J, Pham HT, Schmoellerl J, Javaheri T, Schlederer M, Culig Z, Merkel O, Moriggl R, Grebien F, Kenner L. JAK-STAT signaling in cancer: from cytokines to non-coding genome. Cytokine. 2016;87:26–36. doi: 10.1016/j.cyto.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 32.Prat J. Ovarian carcinomas: five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012;460:237–249. doi: 10.1007/s00428-012-1203-5. [DOI] [PubMed] [Google Scholar]
- 33.Kaldawy A, Segev Y, Lavie O, Auslender R, Sopik V, Narod SA. Low-grade serous ovarian cancer: a review. Gynecol Oncol. 2016;143:433–438. doi: 10.1016/j.ygyno.2016.08.320. [DOI] [PubMed] [Google Scholar]
- 34.Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–1253. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- 35.Liu E, Cote JF, Vuori K. Negative regulation of FAK signaling by SOCS proteins. EMBO J. 2003;22:5036–5046. doi: 10.1093/emboj/cdg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calabrese V, Mallette FA, Deschenes-Simard X, Ramanathan S, Gagnon J, Moores A, Ilangumaran S, Ferbeyre G. SOCS1 links cytokine signaling to p53 and senescence. Mol Cell. 2009;36:754–767. doi: 10.1016/j.molcel.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 37.Souma Y, Nishida T, Serada S, Iwahori K, Takahashi T, Fujimoto M, Ripley B, Nakajima K, Miyazaki Y, Mori M, Doki Y, Sawa Y, Naka T. Antiproliferative effect of SOCS-1 through the suppression of STAT3 and p38 MAPK activation in gastric cancer cells. Int J Cancer. 2012;131:1287–1296. doi: 10.1002/ijc.27350. [DOI] [PubMed] [Google Scholar]
- 38.Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, Harris CC, Herman JG. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 39.Fukushima N, Sato N, Sahin F, Su GH, Hruban RH, Goggins M. Aberrant methylation of suppressor of cytokine signalling-1 (SOCS-1) gene in pancreatic ductal neoplasms. Br J Cancer. 2003;89:338–343. doi: 10.1038/sj.bjc.6601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galm O, Yoshikawa H, Esteller M, Osieka R, Herman JG. SOCS-1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood. 2003;101:2784–2788. doi: 10.1182/blood-2002-06-1735. [DOI] [PubMed] [Google Scholar]
- 41.Oshimo Y, Kuraoka K, Nakayama H, Kitadai Y, Yoshida K, Chayama K, Yasui W. Epigenetic inactivation of SOCS-1 by CpG island hypermethylation in human gastric carcinoma. Int J Cancer. 2004;112:1003–1009. doi: 10.1002/ijc.20521. [DOI] [PubMed] [Google Scholar]
- 42.Iwahori K, Serada S, Fujimoto M, Ripley B, Nomura S, Mizuguchi H, Shimada K, Takahashi T, Kawase I, Kishimoto T, Naka T. SOCS-1 gene delivery cooperates with cisplatin plus pemetrexed to exhibit preclinical antitumor activity against malignant pleural mesothelioma. Int J Cancer. 2013;132:459–471. doi: 10.1002/ijc.27611. [DOI] [PubMed] [Google Scholar]
- 43.Zhou H, Miki R, Eeva M, Fike FM, Seligson D, Yang L, Yoshimura A, Teitell MA, Jamieson CA, Cacalano NA. Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res. 2007;13:2344–2353. doi: 10.1158/1078-0432.CCR-06-2303. [DOI] [PubMed] [Google Scholar]
- 44.Kim MH, Kim MS, Kim W, Kang MA, Cacalano NA, Kang SB, Shin YJ, Jeong JH. Suppressor of cytokine signaling (SOCS) genes are silenced by DNA hypermethylation and histone deacetylation and regulate response to radiotherapy in cervical cancer cells. PLoS One. 2015;10:e0123133. doi: 10.1371/journal.pone.0123133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu X, Zheng J, Guo X, Gao X, Liu H, Tu Y, Zhang Y. Reduced expression of SOCS2 and SOCS6 in hepatocellular carcinoma correlates with aggressive tumor progression and poor prognosis. Mol Cell Biochem. 2013;378:99–106. doi: 10.1007/s11010-013-1599-5. [DOI] [PubMed] [Google Scholar]
- 46.Haffner MC, Petridou B, Peyrat JP, Revillion F, Muller-Holzner E, Daxenbichler G, Marth C, Doppler W. Favorable prognostic value of SOCS2 and IGF-I in breast cancer. BMC Cancer. 2007;7:136. doi: 10.1186/1471-2407-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iglesias-Gato D, Chuan YC, Wikstrom P, Augsten S, Jiang N, Niu Y, Seipel A, Danneman D, Vermeij M, Fernandez-Perez L, Jenster G, Egevad L, Norstedt G, Flores-Morales A. SOCS2 mediates the cross talk between androgen and growth hormone signaling in prostate cancer. Carcinogenesis. 2014;35:24–33. doi: 10.1093/carcin/bgt304. [DOI] [PubMed] [Google Scholar]
- 48.Letellier E, Schmitz M, Baig K, Beaume N, Schwartz C, Frasquilho S, Antunes L, Marcon N, Nazarov PV, Vallar L, Even J, Haan S. Identification of SOCS2 and SOCS6 as biomarkers in human colorectal cancer. Br J Cancer. 2014;111:726–735. doi: 10.1038/bjc.2014.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoefer J, Kern J, Ofer P, Eder IE, Schafer G, Dietrich D, Kristiansen G, Geley S, Rainer J, Gunsilius E, Klocker H, Culig Z, Puhr M. SOCS2 correlates with malignancy and exerts growth-promoting effects in prostate cancer. Endocr Relat Cancer. 2014;21:175–187. doi: 10.1530/ERC-13-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutherland KD, Lindeman GJ, Choong DY, Wittlin S, Brentzell L, Phillips W, Campbell IG, Visvader JE. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726–7733. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- 51.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He B, You L, Uematsu K, Zang K, Xu Z, Lee AY, Costello JF, McCormick F, Jablons DM. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci U S A. 2003;100:14133–14138. doi: 10.1073/pnas.2232790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T, Yamamoto J, Kubo T, Yoshikawa H. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406–6417. doi: 10.1038/sj.onc.1208788. [DOI] [PubMed] [Google Scholar]
- 54.Weber A, Hengge UR, Bardenheuer W, Tischoff I, Sommerer F, Markwarth A, Dietz A, Wittekind C, Tannapfel A. SOCS-3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene. 2005;24:6699–6708. doi: 10.1038/sj.onc.1208818. [DOI] [PubMed] [Google Scholar]
- 55.Puhr M, Santer FR, Neuwirt H, Susani M, Nemeth JA, Hobisch A, Kenner L, Culig Z. Down-regulation of suppressor of cytokine signaling-3 causes prostate cancer cell death through activation of the extrinsic and intrinsic apoptosis pathways. Cancer Res. 2009;69:7375–7384. doi: 10.1158/0008-5472.CAN-09-0806. [DOI] [PubMed] [Google Scholar]
- 56.Evans MK, Yu CR, Lohani A, Mahdi RM, Liu X, Trzeciak AR, Egwuagu CE. Expression of SOCS1 and SOCS3 genes is differentially regulated in breast cancer cells in response to proinflammatory cytokine and growth factor signals. Oncogene. 2007;26:1941–1948. doi: 10.1038/sj.onc.1209993. [DOI] [PubMed] [Google Scholar]
- 57.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi D, Nomoto S, Kodera Y, Fujiwara M, Koike M, Nakayama G, Ohashi N, Nakao A. Suppressor of cytokine signaling 4 detected as a novel gastric cancer suppressor gene using double combination array analysis. World J Surg. 2012;36:362–372. doi: 10.1007/s00268-011-1358-2. [DOI] [PubMed] [Google Scholar]
- 59.Scheitz CJ, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–4139. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasi W, Jiang WG, Sharma A, Mokbel K. Higher expression levels of SOCS 1,3,4,7 are associated with earlier tumour stage and better clinical outcome in human breast cancer. BMC Cancer. 2010;10:178. doi: 10.1186/1471-2407-10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Francipane MG, Eterno V, Spina V, Bini M, Scerrino G, Buscemi G, Gulotta G, Todaro M, Dieli F, De Maria R, Stassi G. Suppressor of cytokine signaling 3 sensitizes anaplastic thyroid cancer to standard chemotherapy. Cancer Res. 2009;69:6141–6148. doi: 10.1158/0008-5472.CAN-09-0994. [DOI] [PubMed] [Google Scholar]
- 62.Hu H, Zhang Q, Chen W, Wu T, Liu S, Li X, Luo B, Zhang T, Yan G, Lu H, Lu Z. MicoRNA-301a promotes pancreatic cancer invasion and metastasis through the JAK/STAT3 signaling pathway by targeting SOCS5. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz121. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 63.Sharma ND, Nickl CK, Kang H, Ornatowski W, Brown R, Ness SA, Loh ML, Mullighan CG, Winter SS, Hunger SP, Cannon JL, Matlawska-Wasowska K. Epigenetic silencing of SOCS5 potentiates JAK-STAT signaling and progression of T-cell acute lymphoblastic leukemia. Cancer Sci. 2019;110:1931–1946. doi: 10.1111/cas.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long J, Jiang C, Liu B, Dai Q, Hua R, Chen C, Zhang B, Li H. Maintenance of stemness by miR-589-5p in hepatocellular carcinoma cells promotes chemoresistance via STAT3 signaling. Cancer Lett. 2018;423:113–126. doi: 10.1016/j.canlet.2017.11.031. [DOI] [PubMed] [Google Scholar]
- 65.Zhang M, Liu S, Chua MS, Li H, Luo D, Wang S, Zhang S, Han B, Sun C. SOCS5 inhibition induces autophagy to impair metastasis in hepatocellular carcinoma cells via the PI3K/Akt/mTOR pathway. Cell Death Dis. 2019;10:612. doi: 10.1038/s41419-019-1856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan D, Wang W, Su J, Zhang Y, Luan B, Rao H, Cheng T, Zhang W, Xiao S, Zhang M, Jiang FN, Sun Z, Jia Z, Zhong WD, Zhu J. SOCS6 functions as a tumor suppressor by inducing apoptosis and inhibiting angiogenesis in human prostate cancer. Curr Cancer Drug Targets. 2018;18:894–904. doi: 10.2174/1568009618666180102101442. [DOI] [PubMed] [Google Scholar]
- 67.Lai RH, Hsiao YW, Wang MJ, Lin HY, Wu CW, Chi CW, Li AF, Jou YS, Chen JY. SOCS6, down-regulated in gastric cancer, inhibits cell proliferation and colony formation. Cancer Lett. 2010;288:75–85. doi: 10.1016/j.canlet.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Pu XM, Xiong Y. kshv-mir-k12-1-5p promotes cell growth and metastasis by targeting SOCS6 in Kaposi’s sarcoma cells. Cancer Manag Res. 2019;11:4985–4995. doi: 10.2147/CMAR.S198411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge D, Gao AC, Zhang Q, Liu S, Xue Y, You Z. LNCaP prostate cancer cells with autocrine interleukin-6 expression are resistant to IL-6-induced neuroendocrine differentiation due to increased expression of suppressors of cytokine signaling. Prostate. 2012;72:1306–1316. doi: 10.1002/pros.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.