Figure 2.
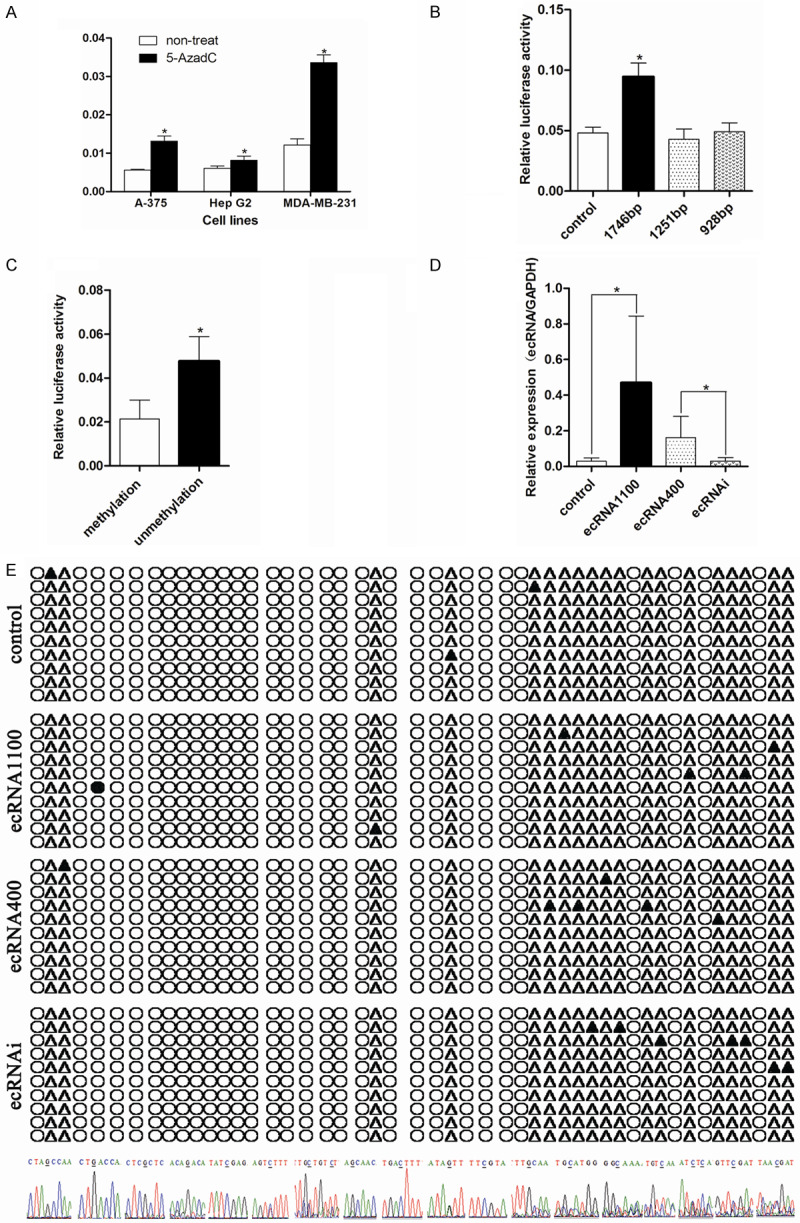
Promoter methylation of gene ARRDC3 was involved in its expression regulation. A. Quantitative analysis of ARRDC3 mRNA in three cell lines before and after treatment with 5-AzadC, the results were expressed as the ratio of copies of the target gene relative to GAPDH. Error bars = SD. The statistics analysis was performed with a t-test and significant differences are indicated by * (P<0.05). ARRDC3 could be inhibited significantly by the reagent 5’-Aza-dC. B. The fluorescence report assay in cell line MDA-MB-231 transfected with the three different length pGL3-ARRDC3 promoter plasmids and pGL3 as control. After statistics analysis with one way ANOVA method, we found the longest promoter plasmid had the maximum transcription activity. C. The fluorescence report assay in cell line MDA-MB-231 transfected with the 1746 bp length pGL3-ARRDC3 promoter plasmid which was methylated or not. The experiment indicated that methylation mediated in the gene ARRDC3 expression. D. The ARRDC3 ecRNA expression level in different stable cell lines detected by real-time quantitative RT-PCR. The statistics analysis was performed with one way ANOVA method and the results demonstrated that these stable cell lines were successfully established. E. Bisulfite sequencing results for different stable cell line (from top to bottom was: control group, ecRNA 1100 group, ecRNA 400 group, and ecRNAi group). Filled circles: methylated CpG sites; open circles: unmethylated CpG sites; filled triangles: methylated CpG sites; open triangles: unmethylated non CpG sites. And the bottom of graph E was the specific sequence representing the methylated sites including the CpG sites or non-CpG sites. We found that ecRNA was not correlated with the methylation of ARRDC3 promoter region.
