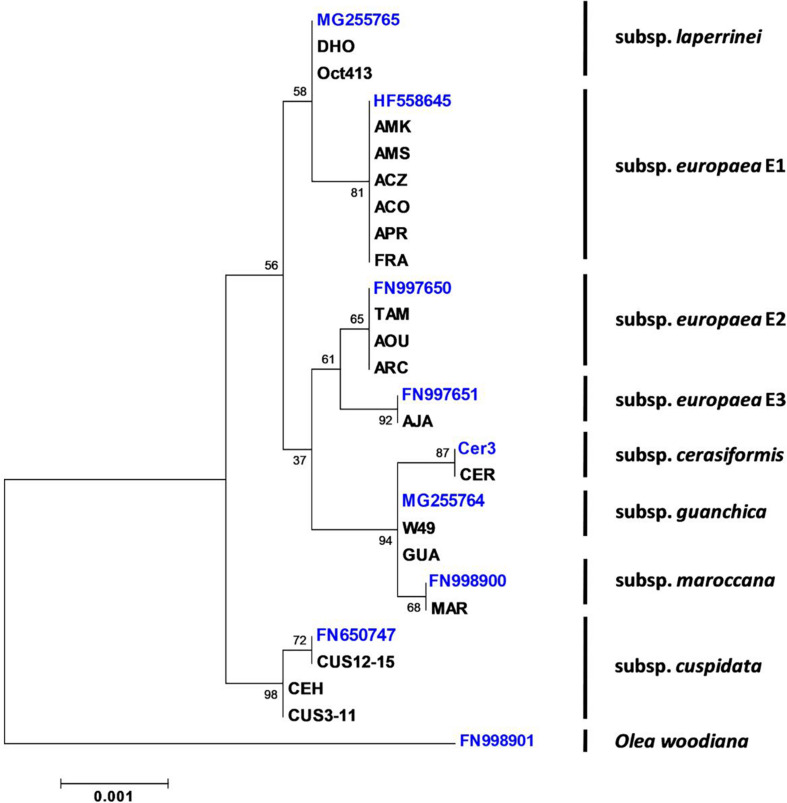
FIGURE 1.
Phylogenetic relationships among wild olive genotypes of the SILVOLIVE collection according to plastid markers. Sequences of the chloroplast markers were used to determine the phylogenetic relationships through the Maximum Likelihood method based on the Tamura 3-parameter model (Tamura, 1992). Phylogenetic analyses were conducted with MEGA6 (Tamura et al., 2013). The tree with the highest log likelihood (–5551.0632) is shown. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 0.05)]. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. The total number of positions in the final dataset was 3,818. Support of nodes was estimated with 1,000 bootstraps. NCBI Accession numbers of the plastomes, used here as reference genomes, are the following: Olea europaea subsp. europaea lineage E1 (HF558645); O. e. subsp. europaea lineage E2 (FN997650); Olea europaea subsp. europaea lineage E3 (FN997651); O. e. subsp. laperrinei (MG255765); O. e. subsp. maroccana (FN998900); O. e. subsp. guanchica (MG255764); O. e. subsp. cuspidata (FN650747); O. woodiana (FN998901). For cerasiformis, with no available full plastome sequence, the mother tree from the CEFE Montpellier collection (Cer3) was used to sequence the chloroplast markers. Other mother trees were also verified: W49 = Olea europaea subsp. guanchica from the WOGB Córdoba collection; Oct413 = Olea europaea subsp. laperrinei variety Dhokar from the WOGB Marrakech collection. Olea woodiana (FN998901) served as the outgroup species to root the tree.