Figure 6:
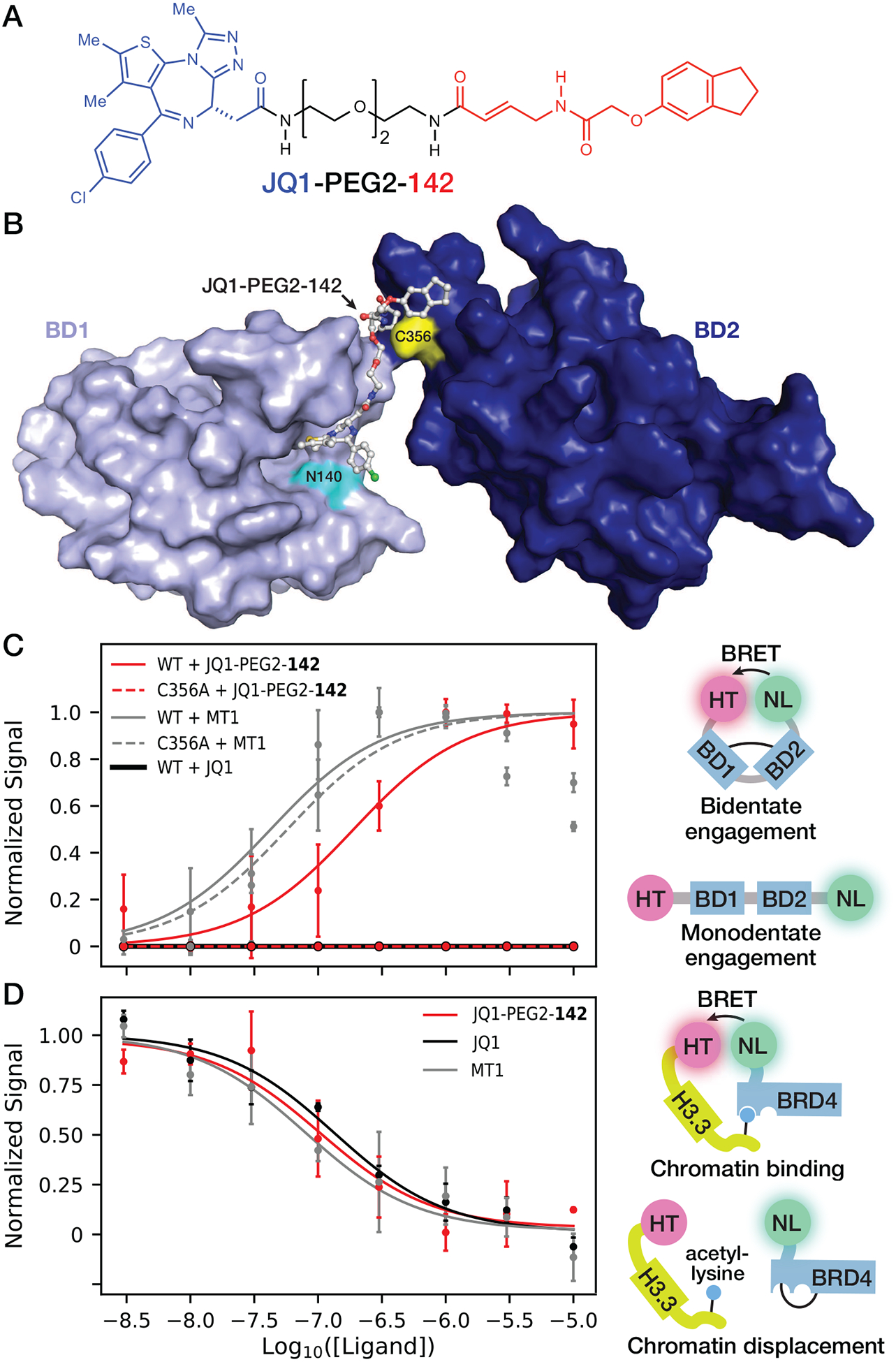
Chemically tethering fragment 142 to JQ1 yields bivalent inhibitors of BRD4 that are active in intact cells. (A) Chemical structure of JQ1 tethered to fragment 142 via a PEG linker. (B) Rosetta model of BRD4-BD1 (PDB ID: 3MXF) docked onto the JQ1 side of JQ1-PEG2-142 after covalent docking to BRD4-BD2 Cys356 (PDB ID: 4KV4) using Schrödinger CovDock (59). (C) NanoBRET assay for bivalent binding to the BRD4 tandem bromodomains. HEK293 cells transfected with either WT or C356A mutant HT-BRD4-BD1/BD2-NL biosensor were treated with 3 nM to 10 μM JQ1-PEG2-142, JQ1 or MT1 and NanoBRET signal was measured as an indicator of conformational change induced by bivalent compound binding. (D) HEK293 cells co-transfected with NL-BRD4 and histone H3.3-HT were titrated with 3 nM to 10 μM JQ1-PEG2-142, JQ1 or MT1 and NanoBRET signal was measured as an indicator of NL-BRD4 displacement from chromatin incorporated with histone H3.3-HT. For NanoBRET experiments, each replicate was normalized according to theoretical minima and maxima of the three-parameter curve fits. Each point represents the average of two or more normalized biological replicates.
