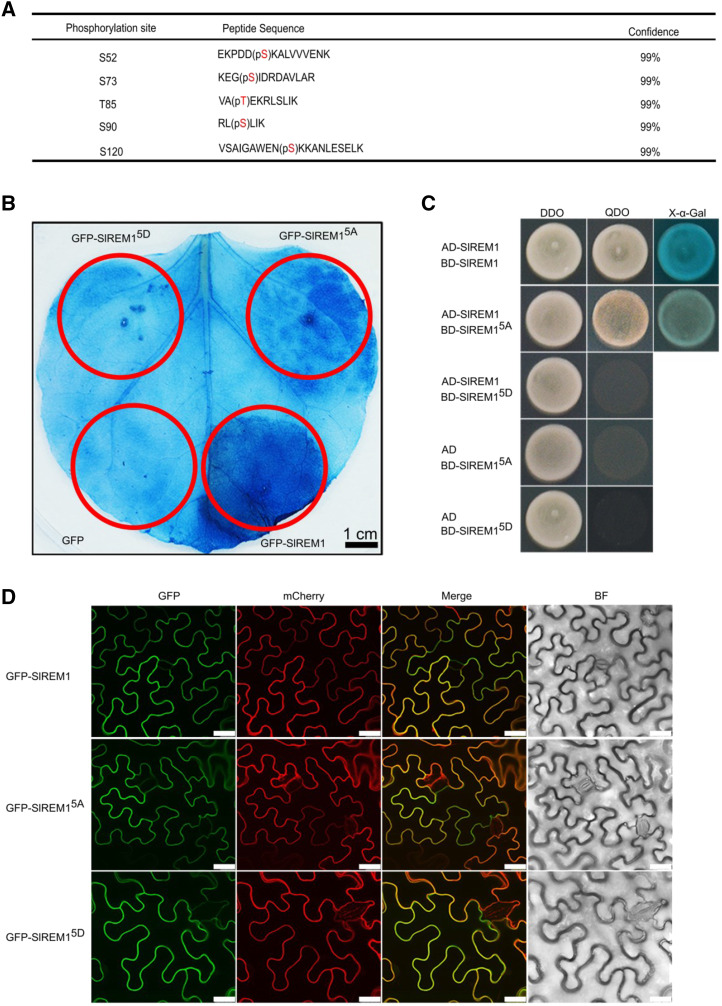
Figure 8.
Five phosphorylation residues are essential for SlREM1-induced cell death. A, Five phosphorylation residues were identified in GFP-SlREM1 and 3×HA-SlREM1 proteins by MS. GFP-SlREM1 and 3×HA-SlREM1 were transiently expressed in N. benthamiana leaves, then purified using anti-GFP and HA beads, respectively, and further analyzed with MS. Red letters within brackets represent the phosphorylated residues. p, Phosphorylation. Phosphorylated peptides were accepted when their confidences calculated by the software ProteinPilot 5.0 were >99%. B, Mutation analysis for the five phosphorylation residues on SlREM1 cell death induction. SlREM15A and SlREM15D indicate that five phosphorylation residues were substituted by Ala or aspartic acids to generate phosphorylation deactivation SlREM15A or activation SlREM15D. Trypan blue staining was performed to evaluate the ability of cell death induction at 5 dpi. The red circles indicate the infiltration areas. C, Y2H analysis of the interactions among SlREM1 and SlREM15A and SlREM15D. DDO, SD/-Leu-Trp agar medium; QDO, SD/-Leu-Trp-His-Ade agar medium; X-α-Gal, 5-Bromo-4-chloro-3-indolyl-α-D-galactopyranoside. D, Mutations in the five phosphorylation residues did not change the localization of SlREM1. GFP-SlREM1, GFP-SlREM15A, and GFP-SlREM15D were transiently expressed in N. benthamiana leaves and the fluorescence was observed at 2 dpi. Scale bars = 25 μm.