Sulfoxidation of NON-RIPENING transcription factor regulates tomato fruit ripening by decreasing its DNA binding capacity and transcriptional regulatory activity.
Abstract
Transcription factors (TFs) are important regulators of plant growth and development and responses to stresses. TFs themselves are also prone to multiple posttranslational modifications (PTMs). However, redox-mediated PTM of TFs in plants remains poorly understood. Here, we established that NON-RIPENING (NOR), a master TF regulating tomato (Solanum lycopersicum) fruit ripening, is a target of the Met sulfoxide reductases A and B, namely E4 and SlMsrB2, respectively, in tomato. Met oxidation in NOR, i.e. sulfoxidation, or mimicking sulfoxidation by mutating Met-138 to Gln, reduces its DNA-binding capacity and transcriptional regulatory activity in vitro. E4 and SlMsrB2 partially repair oxidized NOR and restore its DNA-binding capacity. Transgenic complementation of the nor mutant with NOR partially rescues the ripening defects. However, transformation of nor with NOR-M138Q, containing mimicked Met sulfoxidation, inhibits restoration of the fruit ripening phenotype, and this is associated with the decreased DNA-binding and transcriptional activation of a number of ripening-related genes. Taken together, these observations reveal a PTM mechanism by which Msr-mediated redox modification of NOR regulates the expression of ripening-related genes, thereby influencing tomato fruit ripening. Our report describes how sulfoxidation of TFs regulates developmental processes in plants.
Transcription factors (TFs) are a family of DNA-binding proteins that play vital roles in a wide range of fundamental and disease-related biological processes, including cell cycle and differentiation, development, signaling, stress responses, and cancer development. TFs function by binding to the promoters of target genes to activate or repress their expression in organisms. TFs also undergo posttranslational modifications (PTMs) in response to developmental changes or environmental inputs (Skelly et al., 2016). There are multiple PTM mechanisms in organisms, including acetylation (Zhang et al., 2016), phosphorylation (Kim et al., 2017; Völz et al., 2019), ubiquitination (Zhao et al., 2016; Wu et al., 2017; Zhang et al., 2017), SUMOylation (Zhou et al., 2017; Liu et al., 2019; Nadel et al., 2019), and S-nitrosylation (Kawabe et al., 2018). These PTMs affect protein stability, subcellular localization, interactions with corepressors and activators, and DNA binding activity of TFs, thereby influencing their regulatory activities on target genes.
Recent studies have revealed that redox modification of TFs plays an important role in stress responses or disease-related biological processes. Reactive oxygen species (ROS) are produced in living organisms by aerobic metabolism. During aging and under stress conditions, proteins are susceptible to oxidative damage by ROS. Oxidized proteins and their functions can be partially repaired under certain conditions (Geigenberger et al., 2017; Thormählen et al., 2017). Increasing evidence has shown that oxidative modification of TFs is implicated in regulation of gene expression in response to developmental changes or environmental inputs. The work of Lundquist et al. (2014) showed that redox modification of nuclear actin functions as a regulatory switch to mediate the expression of serum response factor/myocardin-related TF-A (SRF/MRTF-A)-dependent genes. Recently, Wang et al. (2019a) found that oxidation of multiple MiT/TFE TFs links oxidative stress to transcriptional control of autophagy and lysosome biogenesis. In Arabidopsis (Arabidopsis thaliana), oxidative modifications of AtTCP15, AtbZIP16, AtbZIP68, and AtGBF1 significantly decrease their DNA-binding and transcriptional activities (Shaikhali et al., 2012; Viola et al., 2016; Li et al., 2019), whereas oxidation of AtBZR1 leads to enhancement of its transcriptional activity by promoting its interaction with AtARF6 and AtPIF4 (Tian et al., 2018). Interestingly, the study of Tada et al. (2008) demonstrated that thioredoxin-mediated redox modification of NPR1 alters its subcellular localization and regulates disease resistance. These redox modifications are mainly associated with the oxidation of Cys residues in proteins, or the conversion between sulfhydryl and intermolecular/intramolecular disulfide bonds mediated by thioredoxins.
In addition to Cys, Met in proteins is also subject to ROS-dependent oxidative modifications (Levine et al., 2000). Met oxidation in protein, that is, sulfoxidation, leads to the formation of two diastereomers of Met sulfoxide, Met-SO(S) and Met-SO(R), which can be repaired or reversed by the Met sulfoxide reductases MsrA and MsrB, respectively. Previous studies of Msr have mainly focused on its role in aging and oxidative stress (Laugier et al., 2010; Châtelain et al., 2013; Lee et al., 2014). Recent evidence suggests that Msr-mediated sulfoxidation modification is a type of PTM (Valverde et al., 2019) and may modify protein function, thereby regulating various biological processes (Tarrago et al., 2009; Gennaris et al., 2015; Rey and Tarrago, 2018). TFs are important regulatory proteins and it is speculated that Msr-mediated modification of TFs possibly influences the expression of target genes. Unfortunately, few TFs have been confirmed as Msr target proteins, and the biological significance of redox regulation of TFs mediated by Msr is poorly understood.
Fruit is the seed-bearing structure of flowering plants. Fruit ripening is a unique and economically important phase in the life cycle of fruits crops. Tomato (Solanum lycopersicum) is one of the most important commercial crops in the world. Tomato fruit is a good model system to study the mechanisms of fruit ripening due to its simple diploid genetics, well-annotated genome, ease of transformation, short life cycle, and breadth of existing knowledge (Giovannoni et al., 2017). In tomato, NOR is an important ripening regulator, which acts upstream of ethylene synthesis and thereby controls fruit ripening (Barry and Giovannoni, 2007). Our preliminary results from yeast two-hybrid (Y2H) screening for NOR-interacting proteins using a tomato fruit cDNA library identified Msr proteins as interaction partners of NOR. We hypothesized that Msr-mediated sulfoxidation modification of NOR is involved in regulation of fruit ripening in tomato.
In this study, we confirmed that the NOR TF is a target protein of E4, belonging to MsrA family (Supplemental Fig. S1), as well as SlMsrB2 in tomato fruit. Furthermore, mimicking NOR sulfoxidation inhibited tomato fruit ripening by decreasing its DNA-binding capacity and transcriptional regulation of many ripening-related genes. Moreover, NOR sulfoxidation could be partially repaired by E4 and SlMsrB2. These results describe a mechanism of fruit ripening regulation through sulfoxidation of TFs.
RESULTS
Ripening Characteristics of Tomato Fruit Ripening
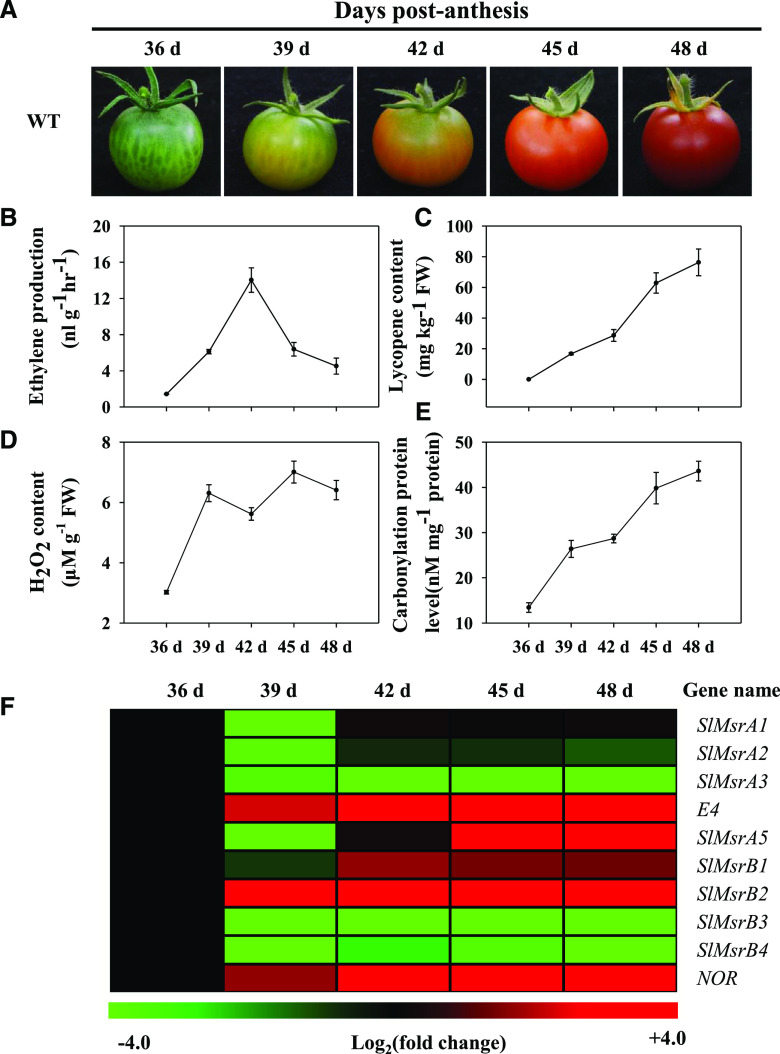
Red appearance of peel is the most important characteristics of tomato fruit ripening. As shown in Figure 1A, tomato fruit turned red at 39 d after anthesis (dpa), accompanied by rapid increases in ethylene production (Fig. 1B) and lycopene accumulation (Fig. 1C). Consistent with the change of fruit color, the accumulation of hydrogen peroxide increased significantly at the peel breaker stage (39 dpa), followed by the rapid increase in protein carbonyl content (Fig. 1, D and E). The results implied that protein oxidation caused by ROS and modification of protein redox status might be related to tomato fruit ripening.
Figure 1.
Ripening characteristics of wild-type (WT) fruit of tomato (S. lycopersicum ‘Ailsa Craig’). A, Ripening phenotype of wild-type tomato fruit. B to E, The changes in ethylene production rate (B), lycopene content (C), H2O2 content (D), and protein carbonyl content (E) during tomato fruit ripening. F, Expression profiles of tomato SlMsrs and NOR genes during fruit ripening, which were determined by RT-qPCR. The expression levels of each gene were expressed as a ratio to the pericarp at 36 dpa, which was set as 1. In the heat map, the values were transformed to log2 (value). Green and red colors indicate down- and up-regulation, respectively. Each value represents the mean ± se of three biological replicates.
Our preliminary Y2H screen revealed interactions between NOR and both E4 and SlMsrB2 in tomato fruits. Therefore, we examined the expression of all SlMsr genes, including five SlMsrAs and four SlMsrBs, as well as the NOR gene, during tomato ripening. Accession numbers of the SlMsr genes and NOR are listed in Supplemental Table S1. As shown in Figure 1F, the transcript levels of SlMsrA1, SlMsrA2, E4, SlMsrA5, SlMsrB1, and SlMsrB2 were up-regulated during fruit ripening and the up-regulations of E4 and SlMsrB2 were much higher than those of other Msr genes. The expression of the NOR gene showed a similar pattern to those of E4 and SlMsrB2 genes. These results suggest that E4 or SlMsrB2 might interact with NOR in vivo to regulate tomato fruit ripening.
NOR Is a Direct Substrate of E4 and SlMsrB2
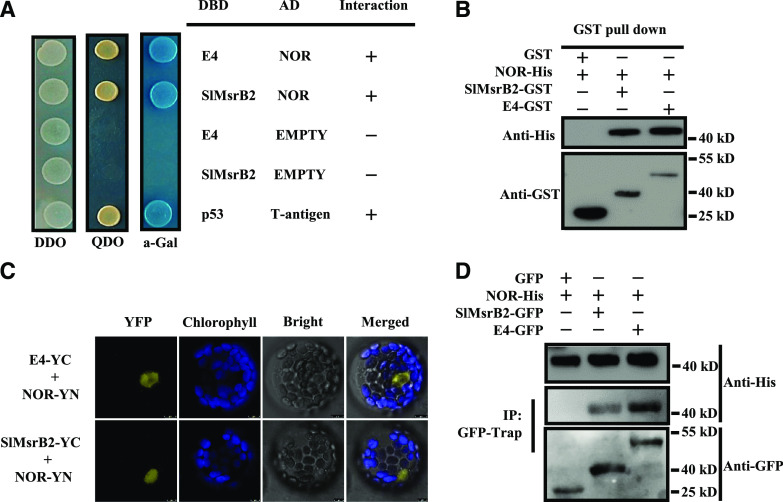
Four methods were applied to verify the interaction between NOR and both E4 and SlMsrB2, including Y2H, bimolecular fluorescence complementation (BiFC), pull-down assay, and coimmunoprecipitation (Co-IP). Y2H analysis showed that the yeast cells cotransformed with DNA-binding domain (DBD)-E4/activation domain (AD)-NOR or DBD-SlMsrB2/AD-NOR grew well on minimal synthetic defined quadruple dropout medium (Fig. 2A), suggesting a physical interaction between NOR and both E4 and SlMsrB2 in vitro. In addition, a pull-down assay showed that GST-E4 or GST-SlMsrB2, but not GST alone, pulled down recombinant His-tagged NOR, also indicating that NOR interacts with E4 and SlMsrB2 in vitro (Fig. 2B). Furthermore, BiFC (Fig. 2C; Supplemental Fig. S2) and Co-IP (Fig. 2D) analyses confirmed the direct interaction between NOR and both E4 and SlMsrB2 in vivo. In addition, subcellular localization indicated that the E4:yellow fluorescent protein (YFP) and SlMsrB2:YFP fusion proteins were predominantly localized to the nucleus (Supplemental Fig. S3), consistent with the nuclear interaction between NOR and both E4 and SlMsrB2 detected by BiFC (Fig. 2C). Taken together, these results demonstrate that NOR physically interacts with E4 and SlMsrB2 in vitro and in vivo.
Figure 2.
NOR interacts with E4 or SlMsrB2 in vitro and in vivo. A, Interaction between NOR and E4 or SlMsrB2 in the Y2H assay. B, In vitro pull-down analysis of NOR- E4/SlMsrB2 interaction. The recombinant GST and GST fusion proteins served as baits, and His-NOR served as prey. C, Interaction between NOR and E4 or SlMsrB2 by bimolecular fluorescence complementation in Arabidopsis mesophyll protoplasts. Yellow signal indicates YFP fluorescence; blue signal indicates chlorophyll autofluorescence. The merged images represent a digital combination of the chlorophyll autofluorescence and YFP fluorescent images. YFP fluorescence was excited at 488 nm, and chlorophyll autofluorescence was excited at 543 nm. Scale bars = 10 µm. D, Interaction between NOR and E4 or SlMsrB2 in the Co-IP assay.
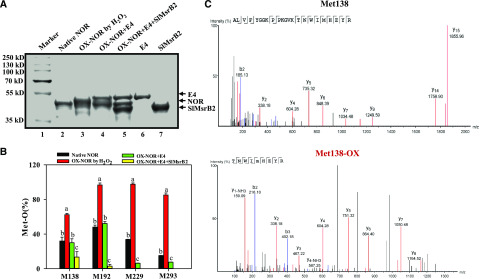
Subsequently, we investigated the effects of E4 and SlMsrB2 on NOR redox status by two different methods, i.e. the gel shift assay and liquid chromatograph tandem mass spectrometry (LC-MS/MS) analysis. Purified NOR was first oxidized by hydrogen dioxide (H2O2). The band representing oxidized NOR was obviously shifted upward in the SDS-PAGE. The oxidized NOR could be effectively reduced by added E4 and SlMsrB2 (Fig. 3A). Then, the oxidized NOR or E4/SlMsrB2-repaired oxidized NOR were subjected to trypsin digestion and LC-MS/MS analysis. Of the eight Met residues in NOR protein, four peptide fragments containing Met-138, Met-192, Met-229, and Met-293, respectively, showed significant decreases in oxidized Met residues after repairing by E4 or SlMsrB2 (Fig. 3B), indicating that these four Met residues were susceptible to redox regulation by Msr. Moreover, E4 and SlMsrB2 seemed involved in the repair of oxidized Met-138 and Met-192, whereas E4 appeared to play a more important role in reducing the oxidized Met-229 and Met-293 (Fig. 3B). These results indicated that E4 and SlMsrB2 are effective reducers of the oxidized NOR protein.
Figure 3.
E4 and SlMsrB2 regulate the redox state of NOR. A, Oxidation of Met in NOR by H2O2 led to a mobility shift of the oxidized protein (OX-NOR).B, E4 and SlMsrB2 can reduce Met-SO in OX-NOR in vitro. The oxidation state of peptides containing Met was determined by LC-MS/MS. Each bar represents the mean ± se of three biological replicates. Different letters above the bars indicated statistically significant differences between the samples (Student’s t test; P < 0.01). C, MS analysis of the tryptic fragments of NOR-GFP protein during tomato fruit ripening. Total protein was extracted from the p35S:NOR-GFP transgenic fruit at 55 dpa, and then incubated with GFP-Trap beads for 4 h. The trapped NOR-GFP was digested with trypsin and then analyzed by LC-MS/MS. The peptide containing Met-138 sulfoxidation is shown highlighted in red. The b- and y-ion series are labeled.
In addition, we analyzed the redox status of NOR in tomato fruit to determine whether NOR is under oxidative modification in vivo. Total protein was extracted from the p35S:NOR-GFP transgenic fruit at 55 dpa, and then incubated with GFP-Trap beads for 4 h. After washing three times with the lysis buffer, the NOR-GFP protein was eluted and digested with trypsin and then analyzed by LC-MS/MS. Two peptide fragments containing the nonoxidized and oxidized Met-138, respectively, were identified (Fig. 3C), suggesting that ROS-induced sulfoxidation occurs on Met-138 in NOR during tomato fruit ripening.
Met Sulfoxidation in NOR Decreases Its DNA Binding Capacity and Transcriptional Regulatory Activity
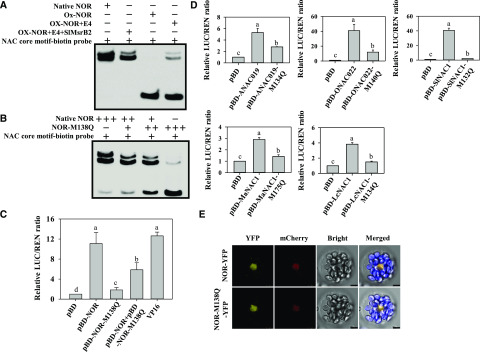
To understand the effect of NOR sulfoxidation modification on its function, the electrophoretic mobility shift assay (EMSA) was used to analyze the DNA binding activity of NOR or oxidized NOR in vitro. As shown in Figure 4A, His-NOR bound strongly to NAM/ATAF1/2/CUC2 (NAC) core motif probes and caused bands to shift upwards, whereas oxidative modification of His-NOR significantly decreased the DNA binding capacity of NOR, which could be partially repaired by adding E4 and SlMsrB2 (Fig. 4A).
Figure 4.
Met sulfoxidation decreases the DNA binding activity and transcription activity of NOR in vitro and in vivo. A, NOR sulfoxidation suppressed the DNA-binding activity of NOR, which were partially restored by E4 and SlMsrB2. B, Mimicking Met-138 sulfoxidation suppressed the DNA-binding activity of NOR. C, Mimicking Met-138 sulfoxidation significantly decreased NOR transactivation activities in vivo. D, Mimicking Met sulfoxidation in conserved M-H-E-Y-R motif significantly decreased the transactivation activities of SlNAC1, ONAC022, ANAC019, LcNAC1, and MaNAC1 in vivo. Each value represents the mean ± se of six biological replicates. Different letters above the bars indicated statistically significant differences between the samples (Student’s t test; P < 0.01). E, Subcellular localization of NOR and NOR-M138Q visualized by YFP analysis. Scale bars = 10 µm.
Tomato NOR protein contains eight Met residues, of which Met-138 is located in the conserved motif M-H-E-Y-R (Supplemental Fig. S4), which has been reported to bind the backbone of the DNA molecule and provide affinity for DNA binding (Welner et al., 2012). As mentioned above, four Met residues, including Met-138, were susceptible to redox regulation by Msr. It is possible that Met-138 sulfoxidation plays an important role in regulating the function of NOR. To verify the speculation, we mutated Met-138 to Gln (Q) to mimic Met sulfoxidation by site-directed mutagenesis (Drazic et al., 2013; Jo et al., 2019) and then evaluated the effect of mimicking Met-138 sulfoxidation on DNA binding capacity and transcriptional regulatory activity of NOR on the reporter gene LUC. The results indicated that mimicked sulfoxidation of Met-138 decreased the DNA binding activity of NOR, which could be partially restored by including a native NOR protein (Fig. 4B). Transient dual-luciferase assays in Nicotiana benthamiana leaves showed that expression of pBD-NOR significantly increased the expression of the LUC reporter in comparison with the expression of the pBD vector alone, and mimicking sulfoxidation in NOR significantly decreased its transcriptional activation activities on LUC (Fig. 4C). Sequence alignment analysis revealed that Met in M-H-E-Y-R motif is conserved in different plant NAC TFs (Supplemental Fig. S4). Arabidopsis ANAC019 (At1g52890), rice (Oryza sativa) ONAC022 (AK107090), tomato SlNAC1 (AY498713), litchi (Litchi chinensis) LcNAC1 (MN650591), and banana (Musa acuminate) MaNAC1 (XP_009406259.1) have been reported to act as transcription activators, respectively (Shan et al., 2014; Zhu et al., 2015; Hong et al., 2016; Meng et al., 2016; Jiang et al., 2017a). We, therefore, mutated the conserved Met in these TFs to Gln, respectively, to mimic Met sulfoxidation and found that the transcriptional activation activities of all the five TFs were significantly decreased (Fig. 4D). These results demonstrate that mimicking sulfoxidation of Met-138 in NOR decreases its DNA binding capacity and transcriptional regulatory activity. Moreover, the decreased transcriptional activity by mimicked sulfoxidation was conserved in different plant NAC TFs.
PTM-mediated alteration of subcellular location of TFs is an important regulatory mechanism of gene expression (Okazaki et al., 2005; Jung et al., 2013). Therefore, we examined whether mimicked sulfoxidation affects the subcellular localization of NOR by analyzing NOR localization in Arabidopsis mesophyll protoplasts by transient expression. The results showed that mimicking sulfoxidation did not affect the subcellular localization of NOR (Fig. 4E).
Mimicking Met-138 Sulfoxidation in NOR Inhibits Tomato Fruit Ripening
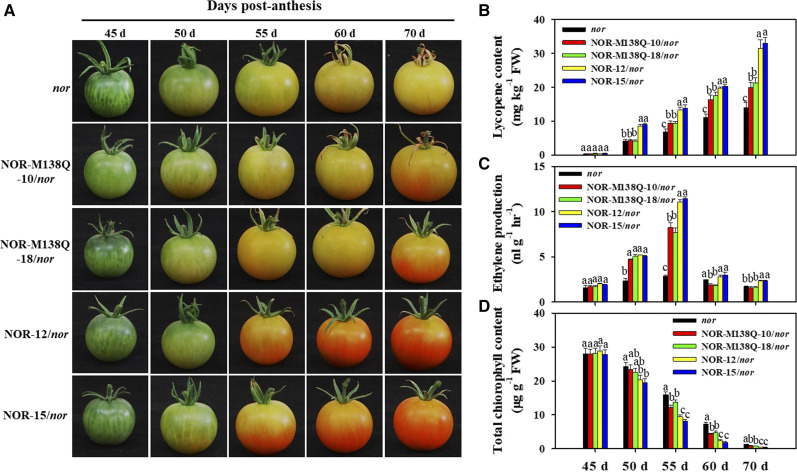
To investigate whether sulfoxidation modification of NOR affects tomato fruit ripening, we generated transgenic tomato plants overexpressing NOR or NOR-M138Q in the nor mutant. Transgenic lines expressing similar protein levels of NOR were selected for phenotypic analysis (Supplemental Fig. S5). As reported previously (Yuan et al., 2016), fruit ripening was inhibited in the nor mutant, which exhibited a failure to turn red and hindered ethylene synthesis and lycopene production (Fig. 5, A–C). The transgenic nor fruits expressing NOR almost completely restored the fruit ripening defects of nor, but the ripening process was delayed (Figs. 1A and 5A). However, the NOR-M138Q transgene only partially rescued the nor fruit ripening phenotype, and was accompanied by decreased ethylene and lycopene synthesis as compared with NOR lines or the nor mutant (Fig. 5, A–C); however, chlorophylls were normally degraded in the fruits of the three genotypes (Fig. 5D). These results indicate that mimicked NOR sulfoxidation represses tomato fruit ripening.
Figure 5.
Mimicking Met-138 sulfoxidation in NOR suppresses tomato fruit ripening. A, Fruit ripening phenotype of nor mutant, NOR, and NOR-M138Q overexpressing in nor lines. Fruit at 45, 50, 55, 60, and 70 dpa from the three genotypes are shown. B to D, The changes in lycopene content (B), ethylene production rate (C), and total chlorophyll content (D) of the three genotypes during fruit ripening. Each value represents the mean ± se of three biological replicates. Different letters above the bars indicated statistically significant differences between the samples (Student’s t test; P < 0.01).
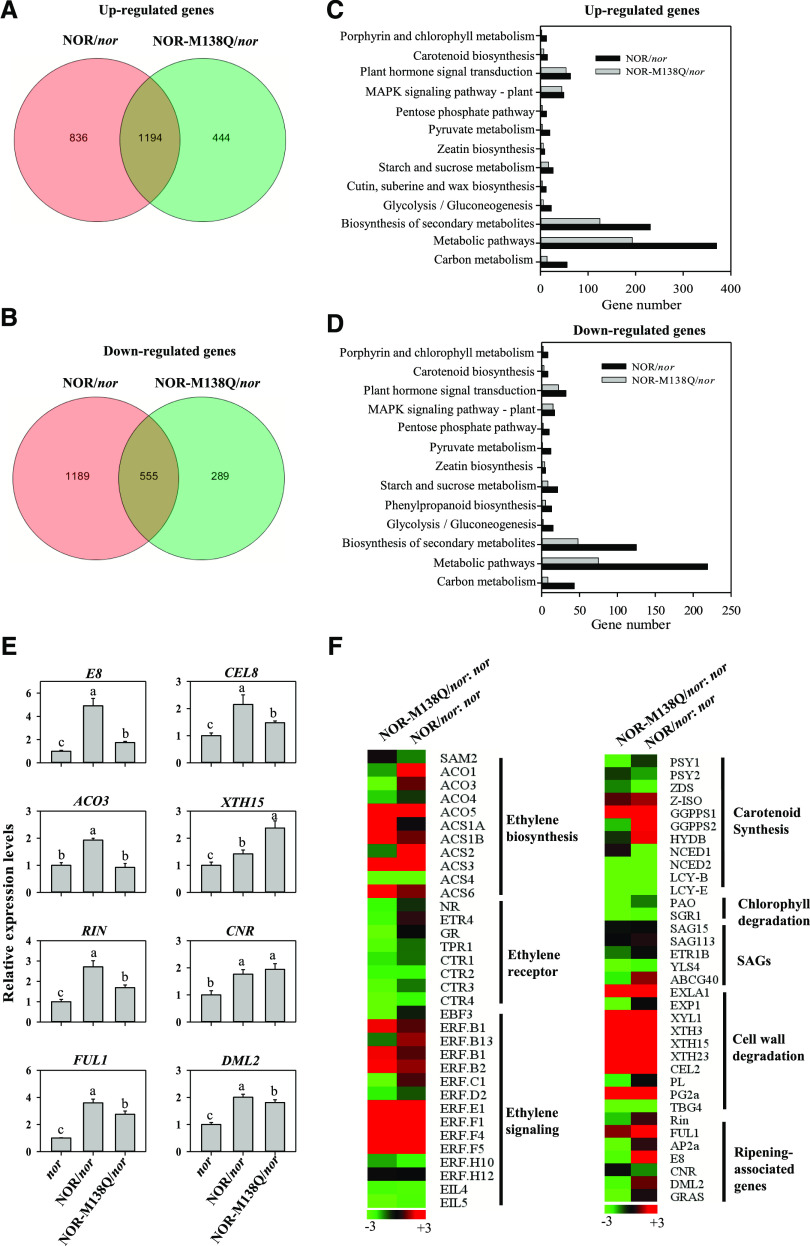
To gain insight into the mechanism by which mimicked NOR sulfoxidation represses tomato fruit ripening, RNA sequencing (RNA-seq) was performed to compare the transcriptomes of the three genotype fruits at 55 dpa, i.e. the nor mutant, the nor mutant complemented with NOR, and the nor mutant transformed with NOR-M138Q. When compared with nor mutant fruit, the expression of 2030 and 1638 genes were up-regulated and that of 1744 and 844 genes were down-regulated in the fruits of the nor mutant expressing NOR or NOR-M138Q, respectively (Fig. 6, A and B; Supplemental Tables S2 and S3). Moreover, the expression of 1,194 genes were coup-regulated, whereas the expression of 555 genes were codown-regulated in the fruits of NOR- and NOR-M138Q-expressing nor mutants (Fig. 6, A and B). GO functional classification analysis showed that the up- and down-regulated genes were mainly enriched in metabolic pathways, biosynthesis of secondary metabolites, plant hormone signaling, and MAPK signaling (Fig. 6, C and D). The reliability of RNA-seq data were confirmed by analyzing the expression levels of 8 genes, including E8, ACO3, RIN, FUL1, CEL8, XTH15, CNR, and DML2 in the fruits of the three genotypes by reverse transcription-quantitative PCR (RT-qPCR; Fig. 6E). Furthermore, many key genes involved in ethylene biosynthesis and signal transduction, brassinosteroid biosynthesis, cutin/suberine/wax biosynthesis, carotenoid biosynthesis, cell wall degradation, and transcriptional regulation were repressed by sulfoxidation of NOR (Fig. 6, C, D, and F; Supplemental Tables S4 and S5). The data suggests that NOR sulfoxidation inhibits tomato fruit ripening by repressing the transcription of ripening-associated genes.
Figure 6.
Mimicking Met-138 sulfoxidation affects the expression of genes related to tomato fruit ripening. A and B, The overlaps of the up-regulated (A) and the down-regulated (B) gens in both the fruits of NOR and NOR-M138Q overexpressing in nor at 55 dpa, compared with nor fruit. C and D, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for the up-regulated (C) and the down-regulated (D) genes in both the fruits of NOR and NOR-M138Q overexpressing in nor at 55 dpa, compared with nor fruit. The y axis indicates differentially expressed genes enriched in KEGG pathways, and the x axis indicates the gene number. E, Validation of RNA-seq results by RT-qPCR. Eight differentially expressed genes were randomly selected to validate by RT-qPCR. Actin gene was used as the internal control. Each bar represents the mean ± se of three biological replicates. Different letters above the bars indicated statistically significant differences between the samples (Student’s t test; P < 0.01). F, Heat map of the expression of ripening-associated genes in the fruits of NOR and NOR-M138Q overexpressing in nor at 55 dpa by RNA-seq.
Mimicked Sulfoxidation of Met-138 in NOR Decreases its DNA Binding Capacity and Transcriptional Activation Activity in Tomato Fruit
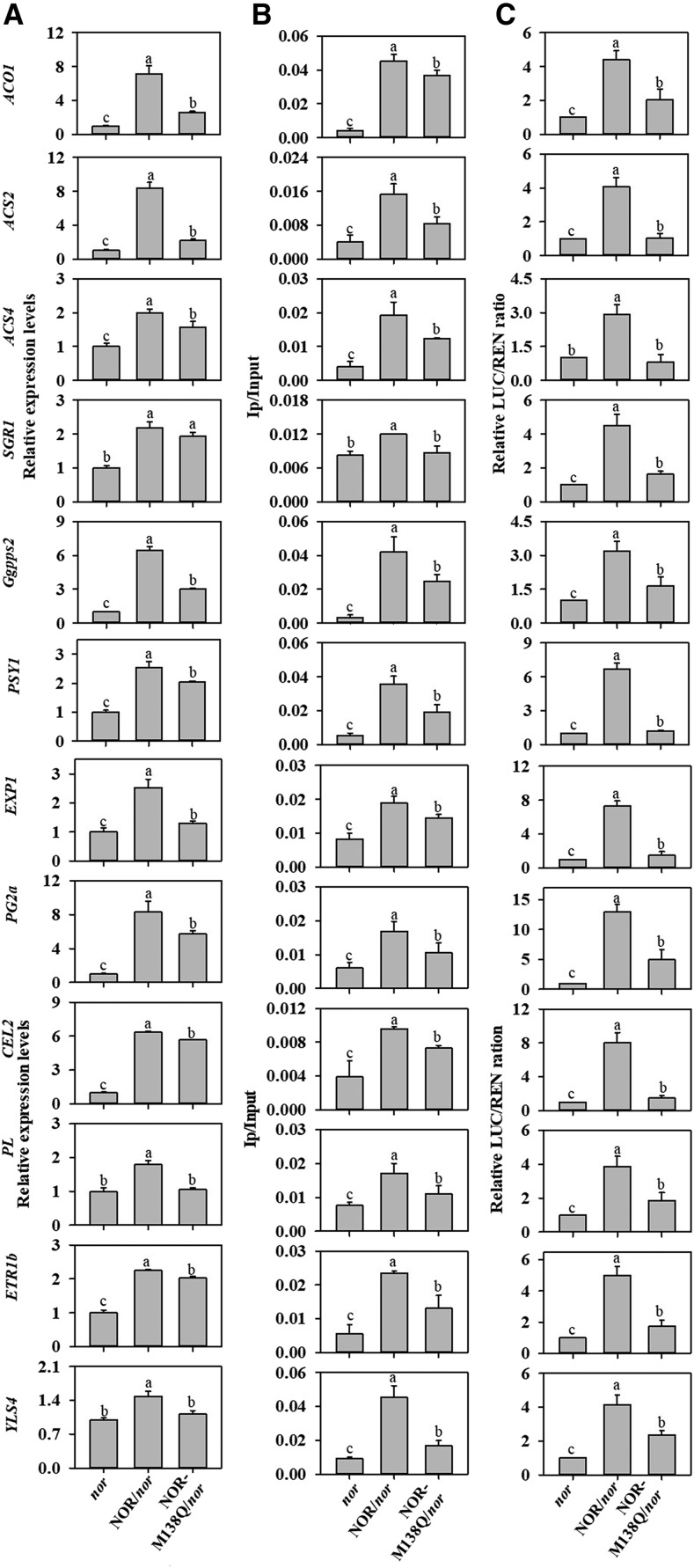
To further elucidate the mechanism that NOR sulfoxidation represses ripening-related genes in tomato fruit, the effects of mimicked NOR sulfoxidation on the DNA binding activities of NOR with the promoters of the key ripening-related target genes and on the transcription activity of the corresponding target genes were examined. Twelve genes including ACO1, ACS2, ACS4, PG2a, PL, CEL2, EXP1, Ggpps2, PSY1, ETR1b, YLS4, and SGR1 were selected from the up-regulated genes in the fruits of the nor mutant complemented with NOR, and the nor mutant transformed with NOR-M138Q, in comparison with the nor mutant fruit. These genes are involved in ethylene biosynthesis, cell wall degradation, carotenoid biosynthesis, chlorophyll degradation, and senescence. RT-qPCR analysis showed that the expression of these ripening-related genes were significantly increased in p35S:NOR fruits at 55 dpa compared with p35S:NOR-M138Q fruits (Fig. 7A), which is consistent with the restored ripening phenotype (Fig. 5A). Chromatin immunoprecipitation (ChIP)-qPCR analysis revealed that NOR transiently interacted with the promoters of these ripening-related genes (Fig. 7B; Supplemental Fig. S6). Mimicking sulfoxidation at Met-138 residues decreased in vivo DNA binding activity of NOR at the promoters of the ripening-related genes. In the transient assays, mimicking sulfoxidation of Met-138 residues also reduced the transcriptional activation activity of NOR (Fig. 7C). Consistent results from RT-qPCR, ChIP-qPCR, and dual-luciferase reporter assay were obtained. Taken together, these results suggest that mimicking sulfoxidation at Met-138 residues in NOR decreases its DNA binding capacity and transcriptional regulatory activity, thereby repressing the expression of ripening-related genes in tomato fruit.
Figure 7.
Met sulfoxidation decreases the DNA binding activity and transcription activity of NOR in tomato fruit. A, The expression levels of 12 ripening-associated genes in the fruits of nor mutant, NOR, and NOR-M138Q overexpressing in nor lines at 55 dpa. B, The binding activities of NOR or NOR-M138Q with the promoters of the 12 genes. C, The transcription activities of the 12 genes by NOR or NOR-M138Q. Each value represents the mean ± se of three biological replicates. Different letters above the bars indicated statistically significant differences between the samples (Student’s t test; P < 0.01).
E4 and SlMsrB2 Are the Direct Targets of NOR
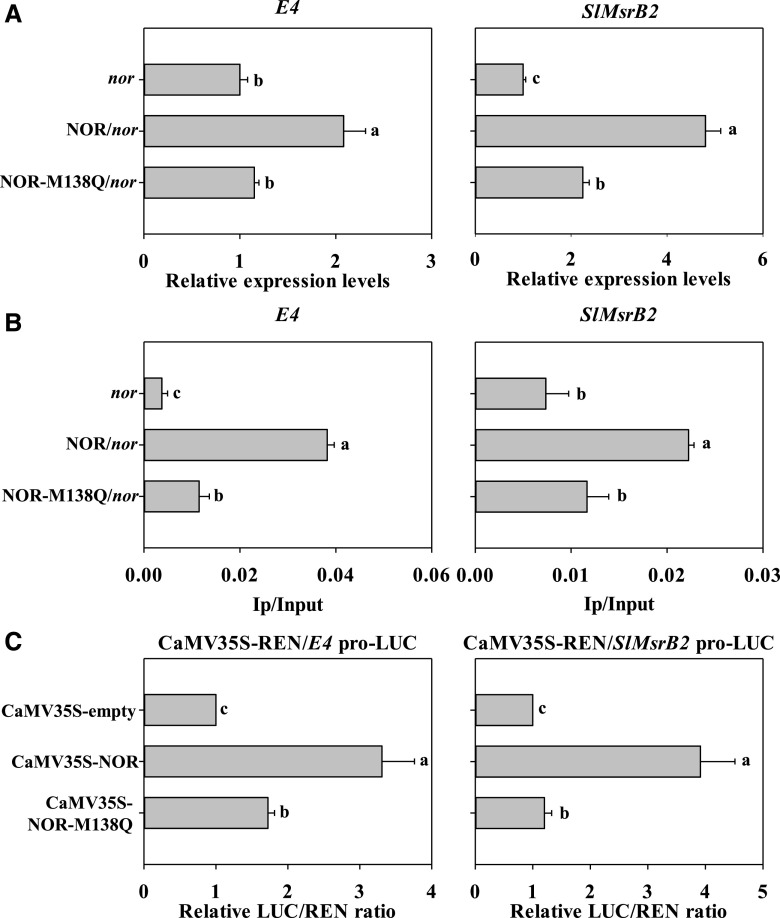
Interestingly, RNA-seq analysis showed that the expression of E4 and SlMsrB2 were up-regulated in the fruit of the nor mutant complemented with NOR when compared with that in the nor mutant fruit (Supplemental Table S2). We speculated that NOR might also directly regulate the expression of E4 and SlMsrB2 in tomato fruit. RT-qPCR analysis indicated that the expression of E4 and SlMsrB2 was increased in the transgenic tomato fruits expressing p35S:NOR-GFP as compared with that in the nor mutant background (Fig. 8A). ChIP-qPCR analysis demonstrated that NOR interacted with the promoters of E4 and SlMsrB2 and mimicking sulfoxidation at Met-138 decreased the interaction between NOR and the promoters of E4 and SlMsrB2 (Fig. 8B). Dual-luciferase reporter assay further confirmed that NOR could activate the promoter activity of E4 and SlMsrB2 in N. benthamiana, but mimicking sulfoxidation decreased the transcriptional activation activity. These results clearly indicate that NOR also activates E4 and SlMsrB2 by directly binding to their promoters.
Figure 8.
Regulation of E4 and SlMsrB2 by TF NOR. A, Expression levels of E4 and SlMsrB2 genes in the fruits of nor mutant, NOR, and NOR-M138Q overexpressing in nor lines at 55 dpa. B, The binding activities of NOR or NOR-M138Q with the promoters of E4 and SlMsrB2. C, The transcription activities of E4 and SlMsrB2 genes by NOR or NOR-M138Q. Each value represents the mean ± se of six biological replicates. Different letters above the bars indicated statistically significant differences between the samples (Student’s t test; P < 0.01).
DISCUSSION
Redox modification of macromolecules is a ubiquitous process in living organisms, and proteins are the main targets of ROS-induced oxidative damage. However, oxidized proteins can be repaired or reversed under certain conditions. Msrs are important redox modification-related enzymes that catalyze the reduction of MetSO in proteins back to Met (Rey and Tarrago, 2018). Previous research suggested that Msr represents an important mechanism of oxidative protein repair and antioxidant defense and plays a role in protecting cells against oxidative damage in animals (Kaya et al., 2015). Overexpression and silencing of Msr by genetic approaches can cause increased or decreased longevity or resistance to oxidative stress in some model organisms (Bruce et al., 2018). Limited information is available on Msrs in relation to their roles in development and responses to stress in plants. Châtelain et al. (2013) reported a positive correlation between Msr capacity and longevity in Arabidopsis seeds. Similarly, fruit senescence is accompanied by down-regulated expression of Msr in litchi (Jiang et al., 2017b). Recently, Jacques et al. (2015) performed a proteome-wide study of Met oxidation in proteins in Arabidopsis upon oxidative stress and identified over 500 sites of oxidation in about 400 proteins. In this study, ROS accumulation (Fig. 1D) and protein carbonylation (Fig. 1E) increased during tomato fruit ripening, indicating that irreversible protein oxidation intensified. Simultaneously, the expression of E4, SlMsrA5, SlMsrB1, and SlMsrB2 genes were obviously up-regulated (Fig. 1F), implying that oxidation of Met residues of proteins, i.e. sulfoxidation, may occur (Fig. 3C). The increased expression of SlMsrs possibly facilitates the reduction of MetSO in proteins back to Met.
Msrs mainly reverse Met sulfoxide back to Met in proteins. Previous studies on Msr functions largely focused on repair of oxidized proteins, which facilitates the resistance to oxidative stress both in vitro and in vivo (Gustavsson et al., 2002; Khor et al., 2004; Laugier et al., 2010; Châtelain et al., 2013; Jacques et al., 2015). A large number of proteins have been confirmed as substrates of Msr in vitro in animals and microorganisms, including calmodulin (CaM; Grimaud et al., 2001), GroEL (a chaperone protein in Escherichia coli.; Khor et al., 2004), Fth (Ezraty et al., 2004), human ether a go-go related gene (hERG) channel (Su et al., 2007), apolipoprotein A-I (Shao et al., 2008), CaMKII (Erickson et al., 2008), TRPM6 channel (Cao et al., 2010), HypT (Drazic et al., 2013), and actin (Lee et al., 2013). However, few Msr target proteins have been identified and characterized in plants for their biological significance (Tarrago et al., 2012; Rey and Tarrago, 2018). Jacques et al. (2015) identified 400 proteins by COFRADIC proteomics method subjected to sulfoxidation in Arabidopsis under oxidative stress, but did not validate the interactions between Msrs and their proteins. Jiang et al. (2017b, 2018) reported that MaCaM1 and LcCaM1 could be target proteins of Msr in banana and litchi fruits, respectively, and are involved in the regulation of fruit ripening and senescence. Recently, Ding et al. (2019) found that MsrA4.1 in wheat (Triticum aestivum) interacts with heme oxygenase 1 to enhance seedling tolerance to salinity or drought stress. In this study, we verified NOR as a substrate of E4 and SlMsrB2 by Y2H, BiFC, pull down, and Co-IP assays (Fig. 2), and found that the oxidized NOR could be repaired by E4 in combination with SlMsrB2 (Fig. 3A). Furthermore, we found that E4 and SlMsrB2 had differential preference for reducing oxidized Met residues in NOR (Fig. 3B). Our data confirms the TF NOR as a target of Msrs in plants.
Recently, Msr-mediated redox modification of functional proteins has emerged as an important PTM mechanism (Valverde et al., 2019). Msrs reversibly modify the redox status of Met residues in proteins susceptible to sulfoxidation, thereby regulating their functions in biological processes. It is well documented that Met oxidation leads to modulation of protein activity or function. Allu et al. (2015) showed that Met sulfoxide reductase MXR2 in Saccharomyces cerevisiae regulates Mge1, a cochaperone of mitochondrial Hsp70, by selectively reducing MetSO at position 155 and restores the activity of Mge1 both in vitro and in vivo. Recently, inactivation of heme oxygenase 1 by Met oxidation has been reported in wheat, whereas the activity is restored by TaMsrA4.1-mediated reduction (Ding et al., 2019). Interestingly, CaMKII, a regulator of calcium flux, is activated by Met oxidation to Met sulfoxide in mice, which is reversed by MsrA (Erickson et al., 2008). Our previous study shows that the mimicked Met oxidation in LcCaM1 does not affect its physical interactions with two LcCaM1-binding senescence-related TFs LcNAC13 and LcWRKY1, but enhances the DNA-binding activities of LcNAC13 and LcWRKY1 (Jiang et al., 2017b). So far, few studies on redox modification of TFs mediated by Msr have been reported. Drazic et al. (2013) showed that mimicked sulfoxidation of HypT, a hypochlorite-responsive TF in E. coli, by substituting Met with Gln, activates HypT activity in regulating its target genes, but the transcriptional activity is inactivated through reduction by MsrA/B. In addition, reversible oxidation of Met 169 in NirA, a fungal nitrate regulator, results in the alteration of subcellular distribution in Aspergillus nidulans (Gallmetzer et al., 2015). In this study, we revealed that Met oxidation in NOR results in the loss of DNA binding capacity, which could be almost completely restored by expression of E4 and SlMsrB2 (Fig. 4A). Moreover, mimicking sulfoxidation of Met-138 in NOR led to an almost complete loss of its DNA binding capacity (Fig. 4B) and transcriptional activation activity (Fig. 4C) and introduction of a native NOR could only partially restore the DNA binding capacity and transcriptional activation activity of NOR-M138Q (Fig. 4, B and C). Furthermore, mimicking sulfoxidation of Met in the conserved M-H-E-Y-R motif of ANAC019, ONAC022, SlNAC1, MaNAC1, and LcNAC1 also resulted in decreased activity of transcriptional activation, suggesting that sulfoxidation regulation of NAC by Msr is conserved in plants. Our results characterize a further PTM of TFs in plants, that is, sulfoxidation, which could regulate gene transcription.
NOR is a NAC family TF, which plays a role in tomato fruit ripening (Gao et al., 2018). NOR acts upstream of ethylene synthesis and thereby controls fruit ripening (Barry and Giovannoni. 2007). Yuan et al. (2016) compared the proteome of the nor mutant and wild type Ailsa Craig (AC) and then found that the nor mutation results in down-regulation of ripening-related proteins, including 1-aminocyclopropane-1-carboxylic acid oxidase (ACO), polygalacturonase 2 (PG2), pectate lyase (PL), phytoene synthase 1 (PSY1), ζ-carotene isomerase (Z-ISO), chalcone synthase 1 (CHS1), and other proteins. Recently, Kumar et al. (2018) found that NAC-NOR mutations in ‘Penjar’ tomato attenuate multiple metabolic processes and prolong the fruit shelf life. Furthermore, Wang et al. (2019b) and Gao et al. (2019) showed that the nor mutant phenotype could not be reconstructed with a CRISPR/Cas9 mutation, suggesting that nor is not a null mutation but a dominant negative allele of NOR. In this study, transgenic complementation of the nor mutant with NOR almost rescued the ripening phenotype of nor (Fig. 5). RNA-seq analysis confirmed that the complementation resulted in up-regulation of some key genes related to ethylene biosynthesis and signaling, carotenoid synthesis, cell wall degradation, and transcription regulation, which can explain why NOR can restore the fruit ripening phenotype of the nor mutant (Fig. 6F). These results further prove that NOR TF regulates tomato fruit ripening.
In tomato fruit, expression of NOR itself is regulated by other TFs during fruit ripening. It was shown that NOR is a direct target of the tomato MADS box TF RIPENING INHIBITOR (RIN; Ito et al., 2008; Fujisawa et al., 2013). SlAREB1 can mediate ABA signaling to activate NOR transcription and ultimately promote ethylene synthesis (Mou et al., 2018). Recently, the study of Ma et al. (2019) showed that SlNAP2, a senescence-controlled NAC TF, acts upstream of NOR to regulate its expression. However, PTMs of TF NOR have not been reported. In this study, we found that TF NOR is subjected to sulfoxidation during fruit ripening (Fig. 3C). The fruit ripening phenotype of nor could not be rescued by NOR-M138Q, the mimicked sulfoxidation (Fig. 5, A–D), demonstrating that mimicking the Met sulfoxide state represses tomato fruit ripening. Furthermore, mimicking sulfoxidation of NOR repressed the expression of numerous key ripening-related genes in tomato fruit (Fig. 7A), which was correlated with the decreased binding of NOR to the promoters of these genes (Fig. 7B) and decreased NOR-mediated transcriptional activation of these genes (Fig. 7C). Therefore, NOR sulfoxidation appears to act as a PTM of TFs involved in regulation of fruit ripening via regulating expression of ripening-related genes.
Interestingly, E4 and SlMsrB2 were determined to be target genes of NOR (Fig. 8B). Mimicked sulfoxidation of NOR also decreased the binding of NOR with the promoters of E4 and SlMsrB2 (Fig. 8B) and NOR-mediated transcriptional activation of these genes (Fig. 8C). Therefore, sulfoxidation of NOR results in repressed expression of E4 and SlMsrB2 in tomato fruit (Fig. 8A), which may in turn further aggravate the sulfoxidation of NOR. The present data indicate that NOR may function as a TF regulating the expression of downstream redox-responsive Msr genes.
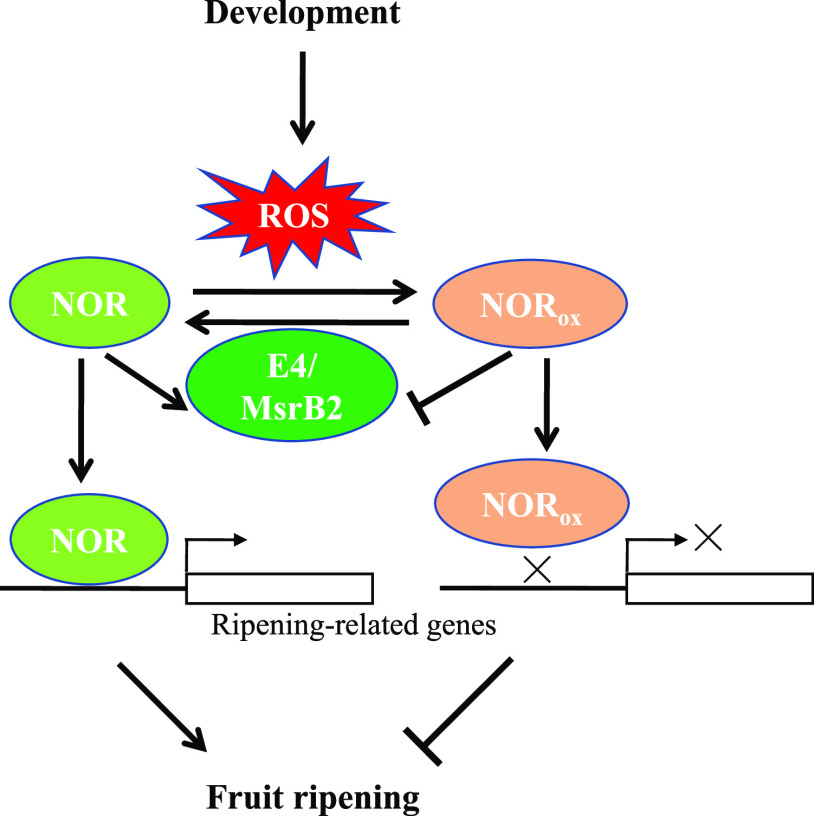
In summary, in this study we characterize a further PTM of TFs in plants, specifically that NOR sulfoxidation regulates the transcription of ripening-related genes and hence fruit ripening in tomato. Met sulfoxidation in NOR leads to decreased transcriptional activity and impaired transcription of ripening-related genes, thereby delaying fruit ripening process in tomato. E4 and SlMsrB2 could partially repair oxidized NOR and restore its transcriptional activity, and thus the proper ripening program can be preserved by Msr enzymes. (Fig. 9).
Figure 9.
A proposed model for the regulatory network involving in redox regulation of NOR by Msr during tomato fruit ripening. During tomato fruit ripening, reactive oxygen species accumulate in large quantities, which lead to protein oxidation, including Met oxidation in NOR (i.e. sulfoxidation). NOR sulfoxidation decreases the binding capacities of NOR with the promoters of ripening-related genes and the transcriptional activities, thereby inhibiting tomato fruit ripening. The oxidized NOR is reduced by E4 and SlMsrB2 to restore its DNA binding capacity and transcriptional activity. Simultaneously, the expression levels of E4 and SlMsrB2 also are regulated by NOR. Therefore, Msr-mediated dynamic modification of NOR redox status is involved in the regulation of tomato fruit ripening.
MATERIALS AND METHODS
Plant Materials
Wild-type tomato (Solanum lycopersicum ‘Ailsa Craig’) and transgenic lines were grown in a greenhouse under controlled temperature and humidity conditions with natural light. Flowers were tagged using anthesis time to determine fruit ripening stages. Fruits of wild-type, nor mutant (kindly provided by Dr. Daqi Fu from China Agricultural University), and NOR transgenic plants were sampled at equivalent ripening stages according to dpa. Peel tissues were immediately collected after harvested, frozen in liquid nitrogen, and stored at −80°C for further analysis.
Generation of Transgenic Tomato Plants
To mimic Met sulfoxidation, the Met-138 residue in NOR was mutated to Gln (Q) by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene). The mutations were verified by DNA sequencing. In order to generate the 35S:NOR-GFP and 35S:NOR-M138Q-GFP construct, the open reading frame of NOR or NOR-M138Q was cloned into pBI-GFP (kindly provided by Dr. Xunchen Liu from South China Botanical Garden, Chinese Academy of Sciences). The destination vectors were sequence confirmed, then transformed into the Agrobacterium tumefaciens strain GV3101, and finally transformed into the nor mutant tomato (S. lycopersicum ‘Ailsa Craig’) as described previously (Fillatti et al., 1987). The presence of the transgene was verified by PCR and DNA sequencing in the T0 and T1 generations. Based on the PCR and DNA sequencing results, 20 positive transgenic plants were identified and used for the expression assays. The expression of NOR in NOR-OE or NOR-M138Q-OE plants was assessed by RT-qPCR. NOR-OE lines 9, 12, and 15 and NOR-M138Q-OE lines 7, 10, and 18 showed dramatic expression up-regulation, and thus were selected to perform Western blots assays.
Fruit Ripening Parameters
To measure the ethylene production rate, fruits were harvested, weighed, and placed in an open environment for 3 h before measurement to avoid wound-induced ethylene during picking. Ethylene production rate was determined using a Hewlett-Packard 5890 series gas chromatograph equipped with a 25-m HP-PLOT Q capillary column (Agilent Technologies) and a flame ionization detector, as described previously (Gao et al., 2018). Peel lycopene was extracted and measured as previously described (Sun et al., 2015) and expressed as milligram of lycopene per gram fresh weight. The carbonyl content of proteins was spectrophotometrically quantified using a carbonyl-specific reagent (2, 4-dinitrophenylhydrazine) as described previously (Jiang et al., 2017b). The H2O2 content was determined by using a hydrogen peroxide assay kit (Nanjing Jiancheng Biochemical Reagent Co.) in accordance with the manufacturer’s instructions. Each sample contained three replicates with five fruit per replicate.
RNA isolation and RT-qPCR Analysis
Total RNA was extracted from pericarp tissues using the Trizol RNA extraction kit (Transgene) followed by reverse transcription using the PrimeScriptTM reagent kit (TaKara) according to the manufacturer’s protocol. The RT-qPCR reactions were carried out in the ABI 7500 Real-Time PCR System (Applied Biosystems) with SYBRPremix Ex TaqTM (Tli RNaseH Plus), ROX plus according to the manufacturer’s protocol. Gene-specific primers were designed with Primer Express software 3.0 (Applied Biosystems) and are listed in Supplemental Table S6. SlACT (Solyc11g005330) was used as the reference gene.
Purification of Recombinant Proteins
The coding sequences of NOR, NOR-M138Q, E4, and SlMsrB2 were inserted into the pET-28a (Novagen) or PGEX-4T-3 vector (Amersham Biosciences) to construct vectors for expression of the respective corresponding recombinant proteins. GST alone, E4-GST, SlMsrB2-GST, and NOR-His fusion proteins were induced and expressed in the Escherichia coli BL21 (DE3) strain. The recombinant proteins were purified with nickel-nitrilotriacetic acid agarose (Qiagen) or glutathione sepharose 4B (GE Healthcare) following the manufacturer’s instruction.
Subcellular Localization Analysis
The coding regions of NOR, E4, or SlMsrB2 without the stop codon were amplified by PCR and subcloned into the pSAT6-EYFP vector. The fusion vectors and control vector were transformed into Arabidopsis mesophyll protoplasts as described previously (Yoo et al., 2007). After 24 to 48 h of incubation at 22°C, the transformed protoplasts were collected for analysis. YFP fluorescence was observed and captured by a laser confocal microscope (Zeiss 510 Meta).
Y2H Assay
To confirm the interactions between NOR and E4 or SlMsrB2, the coding sequences of NOR, E4, and SlMsrB2 were subcloned into the pGBKT7 or pGADT7 vector to fuse the respective proteins with the DBD or AD to create bait and prey constructs. The vectors were then cotransformed into the yeast strain AH109 by the lithium acetate method and grown on DDO medium (minimal media double dropouts, SD medium supplemented with -Leu/-Trp) according to the manufacturer’s protocol (Clontech) for 3 d. Transformed colonies were plated onto the QDO (minimal media quadruple dropouts, SD medium supplemented with -Leu/-Trp/-Ade/-His) medium to test the possible protein-protein interactions. The ability of yeast cells to grow on QDO medium was scored as a positive interaction. The experiments were repeated three times.
BiFC Assay
The coding sequences of NOR, E4, and SlMsrB2 without stop codons were subcloned into pUC-pSPYNE or pUCpSPYCE vectors. The resulting constructs were used for transient assays through a polyethylene glycol transfection of Arabidopsis mesophyll protoplasts, as described previously (Yoo et al., 2007). After 24 to 48 h of incubation at 22°C, the transformed protoplasts were collected for analysis of YFP signals using a fluorescence microscope (Leica SP8 STED 3X).
Pull-Down Assay
Equal volumes of GST protein alone or E4-GST, or SlMsrB2-GST fusion proteins were incubated with the NOR-His fusion protein in 1.0 mL of pull-down buffer at 4°C with gentle rotation for 4 h. GST beads (40 μL; GE Healthcare) were added and then incubated for 4 h. After extensive washing, the eluted proteins were subjected to SDS-PAGE and Western blot analysis. Immunoblotting was performed by using the anti-His (abcam; ab9180) and anti-GST antibodies (abcam; ab19256), and the chemiluminescent signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Coimmunoprecipitation Assay
Nicotiana benthamiana leaves expressing different constructs as indicated were extracted for proteins with the NEB buffer (20 mm Tris-HCl, pH 7.5; 200 mm NaCl; 1 mm EDTA; 0.5% [v/v] Triton X-100; and 1× protease inhibitors; Roche). After centrifugation at 20,000g for 10 min, the supernatant was incubated with GFP-Trap beads (ChromoTek; gta-20) for 4 h. After extensive washing with wash buffer, beads were denatured by boiling with 2 × SDS sample buffer, and then analyzed by SDS-PAGE. Immunoblotting was performed with anti-GFP (abcam; ab290) and anti-His antibodies (abcam; ab9180).
Oxidation and Reduction Assay of NOR
Oxidation and reduction of NOR were carried out as described previously (Jiang et al., 2017b). NOR was oxidized by H2O2 (1 mm) for 3 h at 22°C in 20 mm Tris-HCl buffer (pH 7.5) containing 1 mm diethylenetriaminepentaacetic acid, followed by gel filtration through a NAP-5 Sephadex G-25 column (GE Healthcare) to remove excess H2O2. In vitro repair of oxidized NOR (NORox) was performed by incubating oxidized proteins (2 µm of NORox) with or without purified E4 and SlMsrB2 (2 µm each) in 20 mm Tris-HCl buffer (pH 7.5) containing 10 mm DTT at 22°C for 1 h. The reaction was stopped by adding trifluoroacetic acid and then subjected to SDS-PAGE.
Protein bands corresponding to different redox status were in-gel digested with trypsin (Promega). The resulting peptides were analyzed by LC–MS/MS on a C18 reverse-phase column. Relative abundance of each Met-containing peptide with different redox status was obtained by integration of peak area intensities, taking into account the extracted ion chromatogram of both double- and triple-charged ions.
EMSA
Recombinant His-tagged NOR or NOR-M138Q proteins were prepared as previously described. Oxidation and repair of NOR were performed as described in the previous section. Before the EMSA assay, NOR or NORox proteins were fractionated and concentrated with an 10 KD Amicon Ultra (Millipore) to remove excess DTT. NAC core motif probes (Supplemental Table S6) were labeled using the Pierce DNA 3′-end biotinylation kit (Thermo Fisher Scientific). The ability of NOR to bind the biotin-labeled NAC core motif probes was evaluated as previously described (Jiang et al., 2017b) using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific) determined as previously described.
Dual-Luciferase Reporter Assay
For NOR transcriptional activity assay, the coding sequence of NOR was subcloned into pBD vector to produce the effector construct (pBD-NOR). The double-reporter vector including a firefly luciferase (LUC) driven by a 35S minimal promoter with five repeats of the GAL4-binding element (5 × GAL4), as well as a 35S promoter-driving renilla luciferase (REN) was used as the internal control. To determine the binding activity of NOR to the promoters of ripening-related genes, the promoters were inserted into pGreenII 0800-LUC as reporter plasmids (Hellens et al., 2005). Gene-specific primers were listed in Supplemental Table S6. The constructed effector and reporter plasmids were cotransformed into N. benthamiana leaves by agroinfiltration with Agrobacterium tumefaciens strain GV3101. The activities of LUC and REN luciferase were measured using a Dual-Luciferase Reporter Assay kit (Promega) after 3 d of cotransformation. The analysis was carried out on a Luminoskan Ascent Microplate Luminometer (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. The ratio of LUC to REN was calculated to reflect the final transcriptional activity. At least six biological replicates were assayed for each combination.
RNA-Seq Assays
Total RNAs were extracted from tomato fruits of nor, NOR-12, and NOR-M138Q-18 lines at 55 dpa as described above, and the mRNA-seq libraries were prepared by using the mRNA Seq Kit (Illumina). RNA-seq was performed by Gene Denovo Co. using Illumina HiSeq 2000 with three biological replicates. Clean reads from each sample were aligned to the tomato reference genome (version SL2.50) using TopHat software (version 2.0.14). Fold change ≥ 2, P < 0.05, and false discovery rate <0.05 were set as the significant threshold for differentially expressed genes.
ChIP Assays
ChIP assays were performed as previously described (Kuang et al., 2017). Tomato fruits from NOR-12, NOR-M138Q-18, and the nor mutant at 55 dpa were harvested and immediately cross-linked with 1% (v/v) formaldehyde. After cross-linking, the chromatin from fruit pericarp was extracted and then sheared into an average length of 500 bp by sonication. The chromatin was immunoprecipitated with an anti-GFP (abcam; ab290) antibody. The abundance of immunoprecipitated chromatin was determined by RT-qPCR using the primers given in Supplemental Table S6 and calculated to the relative enrichment relative to the input. RT-qPCR primers were designed to flank the NOR-binding sites within the promoter of potential target genes. The Actin gene was used as a negative control.
Data Handling
Data were expressed as the means ± se. Differences among different treatments were compared using SPSS Version 7.5 (SPSS).
Accession Numbers
Sequence data from this article can be found in the GenBank under the following accession numbers: ANAC019, At1g52890; ONAC022, AK107090; SlNAC1, AY498713; LcNAC1, MN650591; MaNAC1, XP_009406259.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of tomato Msrs with Arabidopsis Msrs.
Supplemental Figure S2. Negative control of BiFC analysis of NOR-E4/SlMsrB2 interaction in vivo.
Supplemental Figure S3. Subcellular localization of E4 and SlMsrB2 visualized by YFP analysis.
Supplemental Figure S4. Protein sequence alignment of NOR with other NAC TFs using Clustal X.
Supplemental Figure S5. Western blot analysis of GFP-fusion protein level from the fruits of nor mutant, NOR/nor, and NOR-M138Q/nor at 55 d. Actin was used as the internal control.
Supplemental Figure S6. Negative control of ChIP-qPCR.
Supplemental Table S1. Gene identification and annotation.
Supplemental Table S2. Differentially expressed genes (DEGs) between NOR/nor and the nor mutant.
Supplemental Table S3. DEGs between NOR-M138Q/nor and the nor mutant.
Supplemental Table S4. KEGG pathway analysis for DEGs between NOR/nor and the nor mutant.
Supplemental Table S5. KEGG pathway analysis for DEGs between NOR-M138Q/nor and the nor mutant.
Supplemental Table S6. Primers used in this study.
Acknowledgments
We thank Daqi Fu (China Agricultural University, Beijing, China) for providing the mutants nor and Xuncheng Liu (South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China) for providing the pBI-GFP vector.
Footnotes
This study was supported by the National Natural Science Foundation of China (grant nos. 31830070, 31871856, 31772041, and 31770726) and the Science and Technology Planning Project of Guangzhou (grant nos. 201804020041 and 201904010014).
References
- Allu PK, Marada A, Boggula Y, Karri S, Krishnamoorthy T, Sepuri NBV(2015) Methionine sulfoxide reductase 2 reversibly regulates Mge1, a cochaperone of mitochondrial Hsp70, during oxidative stress. Mol Biol Cell 26: 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ(2007) Ethylene and fruit ripening. J Plant Growth Regul 26: 143–159 [Google Scholar]
- Bruce L, Singkornrat D, Wilson K, Hausman W, Robbins K, Huang L, Foss K, Binninger D(2018) In vivo effects of methionine sulfoxide reductase deficiency in Drosophila melanogaster. Antioxidants 7: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Lee KP, van der Wijst J, de Graaf M, van der Kemp A, Bindels RJM, Hoenderop JGJ(2010) Methionine sulfoxide reductase B1 (MsrB1) recovers TRPM6 channel activity during oxidative stress. J Biol Chem 285: 26081–26087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Châtelain E, Satour P, Laugier E, Ly Vu B, Payet N, Rey P, Montrichard F(2013) Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc Natl Acad Sci USA 110: 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P, Fang L, Wang G, Li X, Huang S, Gao Y, Zhu J, Xiao L, Tong J, Chen F, Xia G(2019) Wheat methionine sulfoxide reductase A4.1 interacts with heme oxygenase 1 to enhance seedling tolerance to salinity or drought stress. Plant Mol Biol 101: 203–220 [DOI] [PubMed] [Google Scholar]
- Drazic A, Miura H, Peschek J, Le Y, Bach NC, Kriehuber T, Winter J(2013) Methionine oxidation activates a transcription factor in response to oxidative stress. Proc Natl Acad Sci USA 110: 9493–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JR, Joiner MLA, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, et al. (2008) A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133: 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezraty B, Grimaud R, El Hassouni M, Moinier D, Barras F(2004) Methionine sulfoxide reductases protect Ffh from oxidative damages in Escherichia coli. EMBO J 23: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L(1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium-Tumefaciens vector. Nat Biotechnol 5: 726–730 [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y(2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallmetzer A, Silvestrini L, Schinko T, Gesslbauer B, Hortschansky P, Dattenböck C, Muro-Pastor MI, Kungl A, Brakhage AA, Scazzocchio C, et al. (2015) Reversible oxidation of a conserved methionine in the nuclear export sequence determines subcellular distribution and activity of the fungal nitrate regulator NirA. PLoS Genet 11: e1005297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wei W, Zhao X, Tan X, Fan Z, Zhang Y, Jing Y, Meng L, Zhu B, Zhu H, et al. (2018) A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic Res 5: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhu N, Zhu X, Wu M, Jiang CZ, Grierson D, Luo Y, Shen W, Zhong S, Fu DQ, et al. (2019) Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic Res 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Thormählen I, Daloso DM, Fernie AR(2017) The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci 22: 249–262 [DOI] [PubMed] [Google Scholar]
- Gennaris A, Ezraty B, Henry C, Agrebi R, Vergnes A, Oheix E, Bos J, Leverrier P, Espinosa L, Szewczyk J, et al. (2015) Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 528: 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J, Nguyen C, Ampofo B, Zhong S, Fei Z(2017) The epigenome and transcriptional dynamics of fruit ripening. Annu Rev Plant Biol 68: 61–84 [DOI] [PubMed] [Google Scholar]
- Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F(2001) Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem 276: 48915–48920 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Kokke BP, Härndahl U, Silow M, Bechtold U, Poghosyan Z, Murphy D, Boelens WC, Sundby C(2002) A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J 29: 545–553 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA(2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhang H, Huang L, Li D, Song F(2016) Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci 7: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T(2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jacques S, Ghesquière B, De Bock P-J, Demol H, Wahni K, Willems P, Messens J, Van Breusegem F, Gevaert K(2015) Protein methionine sulfoxide dynamics in Arabidopsis thaliana under oxidative stress. Mol Cell Proteomics 14: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Wu F, Li Z, Li T, Gupta VK, Duan X, Jiang Y(2018) Sulfoxidation regulation of Musa acuminata calmodulin (MaCaM) influences the functions of MaCaM-binding proteins. Plant Cell Physiol 59: 1214–1224 [DOI] [PubMed] [Google Scholar]
- Jiang G, Xiao L, Yan H, Zhang D, Wu F, Liu X, Su X, Dong X, Wang J, Duan X, Jiang Y(2017a) Redox regulation of methionine in calmodulin affects the activity levels of senescence-related transcription factors in litchi. Biochim Biophys Acta, Gen Subj 1861(5 Pt A): 1140–1151 [DOI] [PubMed] [Google Scholar]
- Jiang G, Yan H, Wu F, Zhang D, Zeng W, Qu H, Chen F, Tan L, Duan X, Jiang Y(2017b) Litchi fruit LcNAC1 is a target of LcMYC2 and regulator of fruit senescence through its interaction with LcWRKY1. Plant Cell Physiol 58: 1075–1089 [DOI] [PubMed] [Google Scholar]
- Jo I, Kim D, No T, Hong S, Ahn J, Ryu S, Ha NC(2019) Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator. Proc Natl Acad Sci USA 116: 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J(2013) Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci USA 110: 14474–14479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya A, Lee BC, Gladyshev VN(2015) Regulation of protein function by reversible methionine oxidation and the role of selenoprotein MsrB1. Antioxid Redox Signal 23: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe H, Ohtani M, Kurata T, Sakamoto T, Demura T(2018) Protein S-nitrosylation regulates xylem vessel cell differentiation in Arabidopsis. Plant Cell Physiol 59: 17–29 [DOI] [PubMed] [Google Scholar]
- Khor HK, Fisher MT, Schöneich C(2004) Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO-). J Biol Chem 279: 19486–19493 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS(2017) Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res 45: 6613–6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang JF, Chen JY, Liu XC, Han YC, Xiao YY, Shan W, Tang Y, Wu KQ, He JX, Lu WJ(2017) The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol 214: 762–781 [DOI] [PubMed] [Google Scholar]
- Kumar R, Tamboli V, Sharma R, Sreelakshmi Y(2018) NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem 259: 234–244 [DOI] [PubMed] [Google Scholar]
- Laugier E, Tarrago L, Vieira Dos Santos C, Eymery F, Havaux M, Rey P(2010) Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae. Plant J 61: 271–282 [DOI] [PubMed] [Google Scholar]
- Lee BC, Péterfi Z, Hoffmann FW, Moore RE, Kaya A, Avanesov A, Tarrago L, Zhou Y, Weerapana E, Fomenko DE, et al. (2013) MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell 51: 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Li CW, Koh KW, Chuang HY, Chen YR, Lin CS, Chan MT(2014) MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J Exp Bot 65: 5049–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Moskovitz J, Stadtman ER(2000) Oxidation of methionine in proteins: Roles in antioxidant defense and cellular regulation. IUBMB Life 50: 301–307 [DOI] [PubMed] [Google Scholar]
- Li Y, Liu W, Zhong H, Zhang HL, Xia Y(2019) Redox-sensitive bZIP68 plays a role in balancing stress tolerance with growth in Arabidopsis. Plant J 100: 768–783 [DOI] [PubMed] [Google Scholar]
- Liu C, Yu H, Li L(2019) SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet 15: e1007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist MR, Storaska AJ, Liu TC, Larsen SD, Evans T, Neubig RR, Jaffrey SR(2014) Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell 156: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Balazadeh S, Mueller-Roeber B(2019) Tomato fruit ripening factor NOR controls leaf senescence. J Exp Bot 70: 2727–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C, Yang D, Ma X, Zhao W, Liang X, Ma N, Meng Q(2016) Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J Plant Physiol 193: 88–96 [DOI] [PubMed] [Google Scholar]
- Mou W, Li D, Luo Z, Li L, Mao L, Ying T(2018) SlAREB1 transcriptional activation of NOR is involved in abscisic acid-modulated ethylene biosynthesis during tomato fruit ripening. Plant Sci 276: 239–249 [DOI] [PubMed] [Google Scholar]
- Nadel CM, Mackie TD, Gardner RG(2019) Osmolyte accumulation regulates the SUMOylation and inclusion dynamics of the prionogenic Cyc8-Tup1 transcription corepressor. PLoS Genet 15: e1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S, Naganuma A, Kuge S(2005) Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid Redox Signal 7: 327–334 [DOI] [PubMed] [Google Scholar]
- Rey P, Tarrago L(2018) Physiological roles of plant methionine sulfoxide reductases in redox homeostasis and signaling. Antioxidants 7: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikhali J, Norén L, de Dios Barajas-López J, Srivastava V, König J, Sauer UH, Wingsle G, Dietz KJ, Strand Å(2012) Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. J Biol Chem 287: 27510–27525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Lu WJ, Chen JY(2014) Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ 37: 2116–2127 [DOI] [PubMed] [Google Scholar]
- Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW(2008) Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A-I. Proc Natl Acad Sci USA 105: 12224–12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly MJ, Frungillo L, Spoel SH(2016) Transcriptional regulation by complex interplay between post-translational modifications. Curr Opin Plant Biol 33: 126–132 [DOI] [PubMed] [Google Scholar]
- Su Z, Limberis J, Martin RL, Xu R, Kolbe K, Heinemann SH, Hoshi T, Cox BF, Gintant GA(2007) Functional consequences of methionine oxidation of hERG potassium channels. Biochem Pharmacol 74: 702–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhang N, Wang J, Zhang H, Li D, Shi J, Li R, Weeda S, Zhao B, Ren S, et al. (2015) Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J Exp Bot 66: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X(2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrago L, Kieffer-Jaquinod S, Lamant T, Marcellin MN, Garin JR, Rouhier N, Rey P(2012) Affinity chromatography: A valuable strategy to isolate substrates of methionine sulfoxide reductases? Antioxid Redox Signal 16: 79–84 [DOI] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Rey P(2009) Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: Gene organization, reduction mechanisms, and physiological roles. Mol Plant 2: 202–217 [DOI] [PubMed] [Google Scholar]
- Thormählen I, Zupok A, Rescher J, Leger J, Weissenberger S, Groysman J, Orwat A, Chatel-Innocenti G, Issakidis-Bourguet E, Armbruster U, et al. (2017) Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol Plant 10: 168–182 [DOI] [PubMed] [Google Scholar]
- Tian Y, Fan M, Qin Z, Lv H, Wang M, Zhang Z, Zhou W, Zhao N, Li X, Han C, et al. (2018) Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat Commun 9: 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde H, Cantón FR, Aledo JC(2019) MetOSite: An integrated resource for the study of methionine residues sulfoxidation. Bioinformatics 35: 4849–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Camoirano A, Gonzalez DH(2016) Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol 170: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völz R, Rayapuram N, Hirt H(2019) Phosphorylation regulates the activity of INDETERMINATE-DOMAIN (IDD/BIRD) proteins in response to diverse environmental conditions. Plant Signal Behav 14: e1642037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang N, Xu D, Ma Q, Chen Y, Xu S, Xia Q, Zhang Y, Prehn JHM, Wang G, et al. (2019a) Oxidation of multiple MiT/TFE transcription factors links oxidative stress to transcriptional control of autophagy and lysosome biogenesis. Autophagy doi:10.1080/15548627.2019.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tavano ECDR, Lammers M, Martinelli AP, Angenent GC, de Maagd RA(2019b) Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep 9: 1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner DH, Lindemose S, Grossmann JG, Møllegaard NE, Olsen AN, Helgstrand C, Skriver K, Lo Leggio L(2012) DNA binding by the plant-specific NAC transcription factors in crystal and solution: A firm link to WRKY and GCM transcription factors. Biochem J 444: 395–404 [DOI] [PubMed] [Google Scholar]
- Wu MY, Lin CY, Tseng HY, Hsu FM, Chen PY, Kao CF(2017) H2B ubiquitylation and the histone chaperone Asf1 cooperatively mediate the formation and maintenance of heterochromatin silencing. Nucleic Acids Res 45: 8225–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan X-Y, Wang R-H, Zhao X-D, Luo Y-B, Fu D-Q(2016) Role of the tomato non-ripening mutation in regulating fruit quality elucidated using iTRAQ protein profile analysis. PLoS One 11: e0164335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Qi B, Wang L, Zhao B, Rode S, Riggan ND, Ecker JR, Qiao H(2016) EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat Commun 7: 13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Li F, Liu H, Yang W, Chong K, Xu Y(2017) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell 43: 731–743 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Ren YR, Wang QJ, Wang XF, You CX, Hao YJ(2016) Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol 172: 1973–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LJ, Li YY, Zhang RF, Zhang CL, Xie XB, Zhao C, Hao YJ(2017) The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ 40: 2068–2080 [DOI] [PubMed] [Google Scholar]
- Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B(2015) Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J 84: 597–610 [DOI] [PubMed] [Google Scholar]