Figure 1.
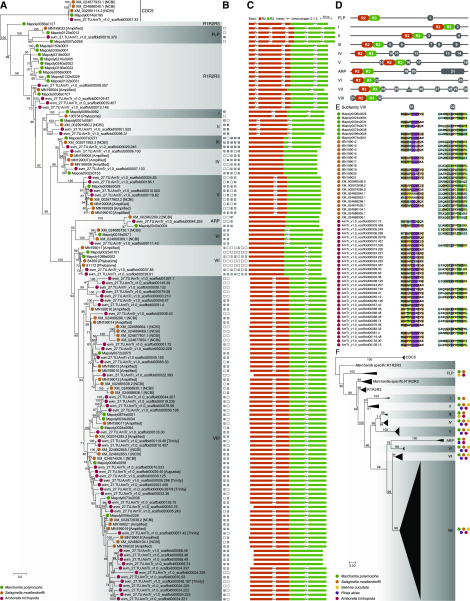
Phylogenetic analysis of proteins with R2R3-MYB domains in representative land plants. A, Maximum likelihood (ML) phylogeny of proteins with R2R3-MYB domains in M. polymorpha, S. moellendorffii, and A. trichopoda. The MYB protein CDC5 sequences were used as the outgroup. Ultrafast bootstrap values are associated with the internal branches; values <50 were omitted. R1R2R3-MYB TFs and 10 subfamilies of R2R3-MYB TFs are labeled. The bracketed text indicates the source database or the sequence amplified in this study. B, The presence or absence of subfamily-specific auxiliary motifs of R2R3-MYB TFs is indicated by a solid or open square, respectively, with number labels corresponding to the terminal nodes of the phylogenetic tree. C, Intron-exon structure of MYB genes in the R2R3-MYB-encoding region. D, Subfamily-specific amino acid auxiliary motifs (gray boxes) of land plant R2R3-MYB TFs. The diagrams are not drawn to scale. E, Subfamily VIII R2R3-MYB protein sequences of motifs 31 (left column) and 32 (right column). F, ML phylogeny of proteins with R2R3-MYB domains in five representative species. R1R2R3-MYB TFs and 10 subfamilies of R2R3-MYB TFs are labeled and collapsed into triangles. Ultrafast bootstrap values are associated with the internal branches. The color of the solid circles in A, E, and F indicates the source of the sequence: green, bryophytes; orange, lycophytes; yellow, ferns; purple, gymnosperms; and red, angiosperms.