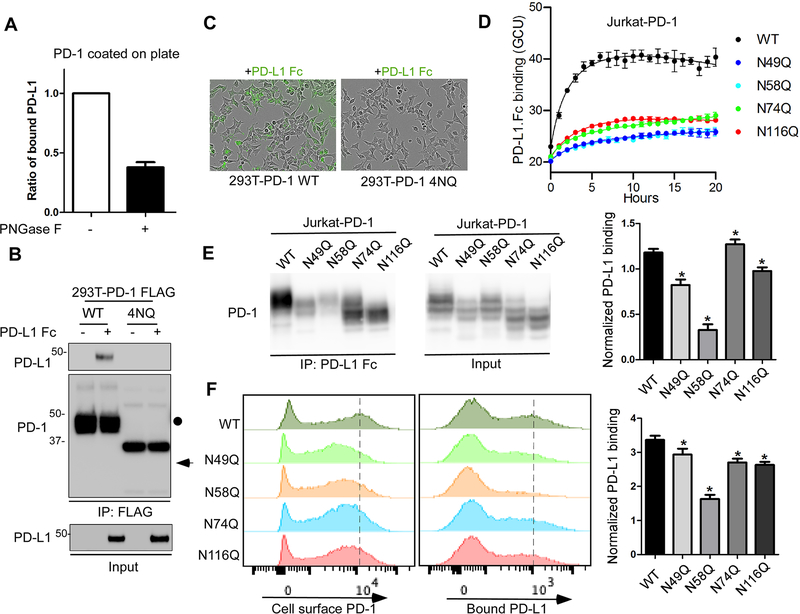
Figure 4. The glycosylation of PD-1 is essential for its interaction with PD-L1.
(A) In vitro plate-based binding analysis of purified PD-1 with PD-L1 Fc in the absence or presence of PNGase F. (B) Analysis of PD-1/PD-L1 binding by immunoprecipitation. The lysates of 293T cells overexpressing FLAG-tagged PD-1 WT or 4NQ mutant were incubated with PD-L1 Fc and anti-FLAG M2 agarose, and then subjected to IP-Western. (C) Time-lapse microscopy of the dynamic interaction between green fluorescent-labeled PD-L1-Fc and PD-1 WT or 4NQ expressed in 293T cells. (D) Time-lapse evaluation and quantitation of the dynamic interaction between green fluorescent-labeled PD-L1-Fc and PD-1 WT or the indicated mutants expressed in Jurkat cells. (E) Mapping of the glycosylated sites critical for binding with PD-L1 by immunoprecipitation. The lysates of Jurkat cells overexpressing PD-1 WT or the indicated NQ mutants were incubated with pan mouse IgG dynabeads and Fc (mouse IgG) fused human PD-L1, and then subjected to IP-Western. The band intensity was quantified and normalized to show PD-L1 binding levels (right panel). (F) Flow cytometric analysis of the binding of PD-L1 Fc with cell surface PD-1 by using Jurkat T cells expressing PD-1 WT or the indicated NQ mutants. Median Fluorescence Intensity was quantified and normalized to show PD-L1 binding levels (right panel). All data represent mean ± SD from at least three independent experiments. *, P < 0.05.