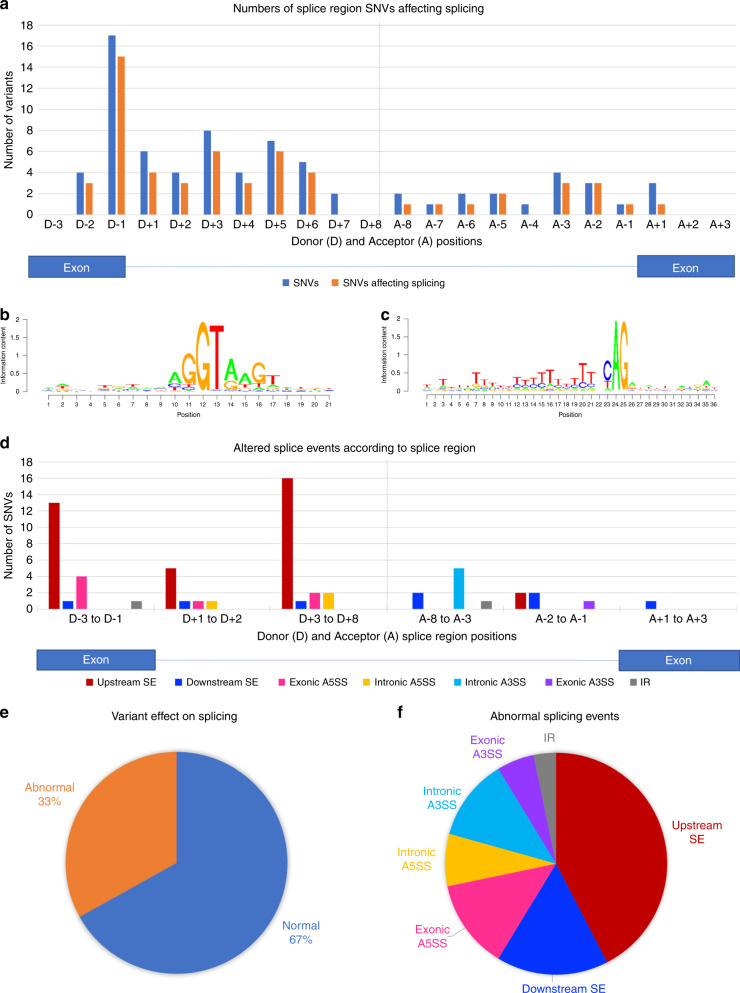
Fig. 1. Variant locations and effects on splicing.
(a) Plot of the numbers of single-nucleotide variants (SNVs) in this cohort (multinucleotide variants not included) present at each donor (D-3 to D+8) and acceptor (A-8 to A+3) splice region position, along with the numbers of these found to affect splicing. (b, c) Position–weight matrices of nucleotide sequence across the splice donor (b) and acceptor (c) regions as determined for the specific exon–intron junctions analyzed in this study. In this representation, the donor splice site +1 position correlates to position 12 in (b), while the acceptor splice site −1 position correlates to position 25 in (c). (d) Abnormal splicing effects plotted by SNV location. Sequence ontology defines the donor splice region as extending from the third last nucleotide of the exon (D-3) to the eighth nucleotide of the intron (D+8) and the acceptor splice region as extending form the eighth last nucleotide of the intron (A-8) to the third nucleotide of the exon (A+3).22 (e) Overall proportion of all variants affecting splicing in this cohort. (f) Proportions of different abnormal splicing events identified in this cohort. A3SS alternative 3´ splice site; A5SS alternative 5´ splice site, IR intron retention, SE skipped exon.