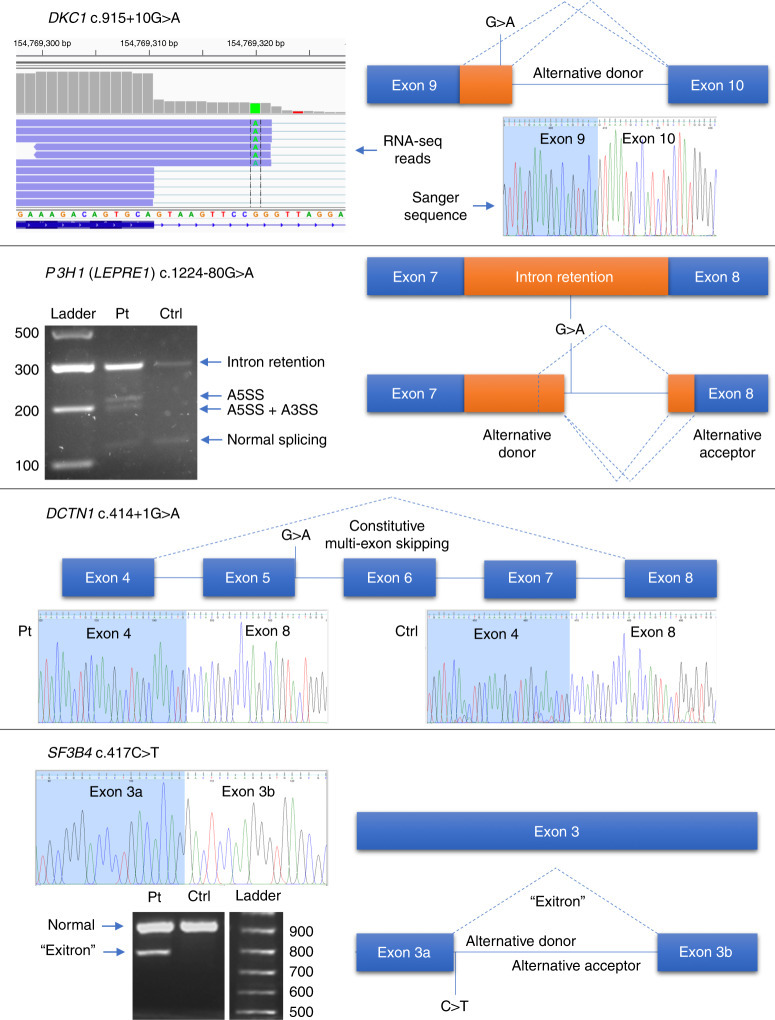
Fig. 2. Illustrative examples of variant splicing analysis.
DKC1 c.915+10G>A could not be identified by reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing but alternative donor splice site usage was identified by RNA-seq. P3H1 (LEPRE1) c.1224-80G>A causes at least three abnormal splicing events using alternative splice donor and acceptor sites, as well as increasing levels of intron retention. DCTN1 c.414+1G>A appears to alter a canonical splice donor site but exons 5–7, although annotated, are never expressed and are constitutively spliced out. SF3B4 c.417C>T is a synonymous coding variant but causes formation of a 125-nt “exitron,” an intronic region within an exon. A3SS alternative 3´splice site, A5SS alternative 5´ splice site, Ctrl control, Pt patient.