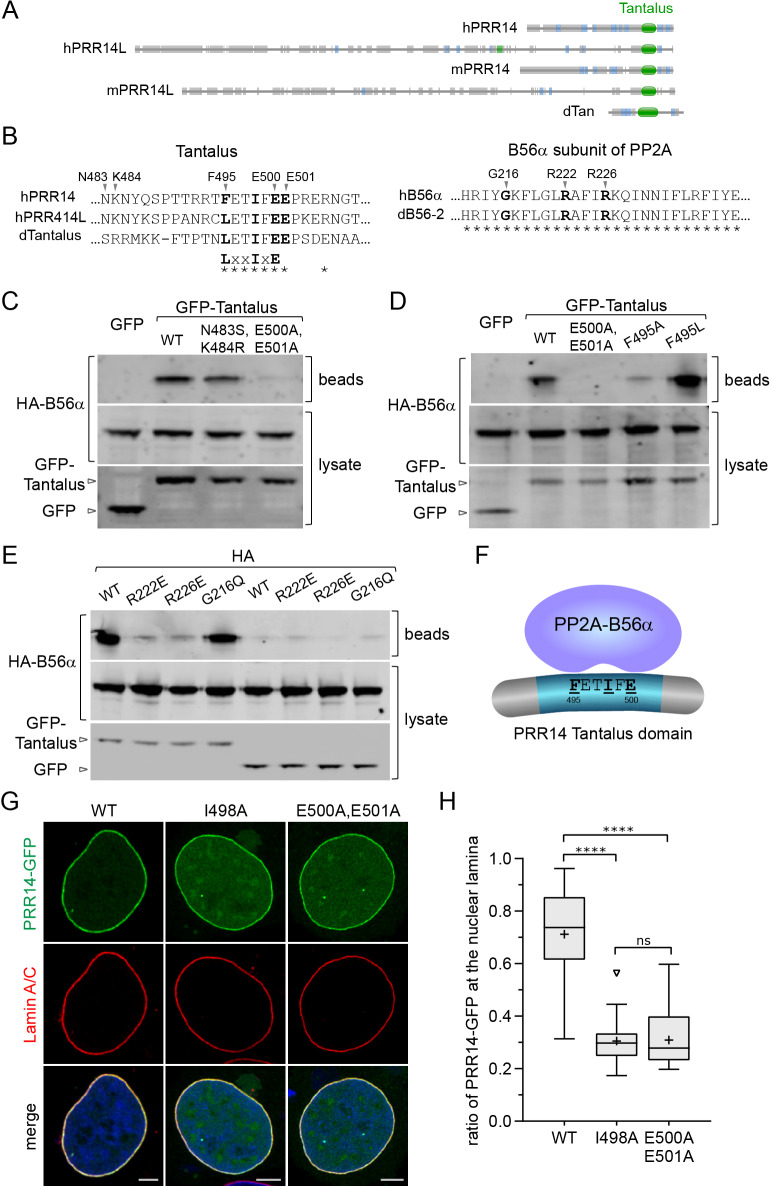
Fig. 7.
PP2A binds PRR14 and regulates PRR14–nuclear lamina association. (A) Diagrams of indicated proteins showing Tantalus domain location. (B) Summary of key residues in the PP2A B56α subunit and PRR14 Tantalus domain that were predicted to mediate B56α binding to the corresponding PRR14 [L/F/M]xxIxE motif. Amino acid sequence identity (*) is indicated. (C) Anti-GFP beads were added to lysates of HeLa cells that had been co-transfected with HA-tagged B56α, and free GFP or GFP fused to the PRR14 Tantalus domain. The indicated Tantalus substitutions were tested for effects on B56α interactions, alongside the wild type (WT). Bead-bound proteins were analyzed by western blotting using anti-HA antibodies to detect B56α–PRR14 Tantalus interactions. Anti-GFP was used to monitor GFP/GFP-Tantalus in the lysates. The E500A, E501A substitution inhibited the interaction as predicted. (D) Using an experimental design as described in C, PRR14 Tantalus domain substitutions E500A, E501A and F495A were found to inhibit binding, whereas F495L increased binding. (E) Using an experimental design as described in C, HA-tagged B56α subunits harboring the indicated amino acid substitutions were assayed for binding to the GFP–Tantalus domain. (F) A schematic diagram showing PP2A interaction with the PRR14 Tantalus domain. (G) Representative confocal images of HeLa cells transfected with the indicated Tantalus domain amino acid substitutions introduced into full-length GFP-tagged human PRR14 (see panel B). The indicated substitutions resulted in reduced nuclear lamina association. (H) Box plot demonstrating the proportion of the indicated PRR14–GFP proteins, as depicted in G, at the nuclear lamina compared to the total nuclear signal, calculated using Lamin A/C signal as a mask. Boxes indicate the interquartile range with the median represented by a horizontal bar. Whiskers are drawn using the Tukey method and + indicates the mean value. n=20 cells per condition. ****P<0.0001; ns, not significant (one-way ANOVA with Dunn's multiple comparison test). Scale bars: 5 µm. Data shown in C–E are representative of more than one experiment.