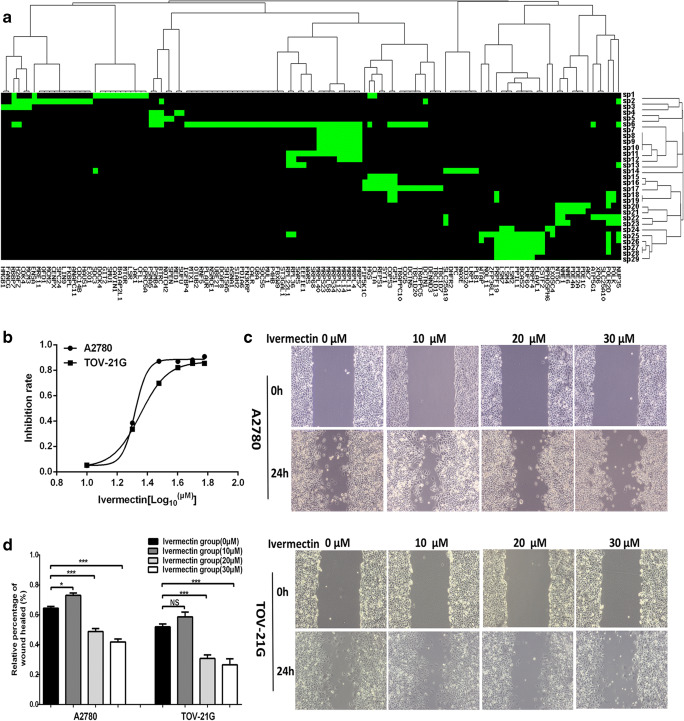
Fig. 1.
Signaling pathways enriched with EIF4A3-binding mRNAs and effects of ivermectin on ovarian cancer cell migration. a Signaling pathways enriched with EIF4A3-binding mRNAs were clustered into 8 groups. sp = signaling pathway. sp1 = EGFR1; sp2 = Cell cycle; sp3 = Retinoblastoma gene in cancer; sp4 = Hedgehog; sp5 = Notch; sp6 = Metabolism of proteins; sp7 = Mitochondrial translation; sp8 = Mitochondrial translation elongation; sp9 = Mitochondrial translation initiation; sp10 = Mitochondrial translation termination; sp11 = Translation; sp12 = Ribosome; sp13 = Selenoamino acid metabolism; sp14 = Metabolism of proteins; sp15 = Cargo recognition for clathrin-mediated endocytosis; sp16 = Clathrin-mediated endocytosis; sp17 = Membrane trafficking; sp18 = Nucleotide excision repair; sp19 = Transcription coupled nucleotide excision repair (TC-NER); sp20 = Purine metabolism; sp21 = Purine metabolism Homo sapiens (human); sp22 = Purine nucleotides nucleosides metabolism; sp23 = Pyrimidine metabolism; sp24 = RNA degradation Homo sapiens (human); sp25 = Metabolism of RNA; sp26 = Processing of capped intron-containing pre-mRNA; sp27 = mRNA splicing; sp28 = mRNA splicing - major pathway; and sp29 = Spliceosome Homo sapiens (human). b Cell viability was measured with CCK8 assay in OC cells A2780 and TOV-21G treated with ivermectin (0–60 μM) for 24 h (n = 3; X = Log (ivermectin concentration)). c Wound healing test of OC cells A2780 and TOV-21G treated with ivermectin (0–30 μM) for 24 h (n = 3). d Histogram statistics of wound healing test of OC cells A2780 and TOV-21G treated with ivermectin (0–30 μM) (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001