Abstract
Legionella pneumophila (Lp) is a water borne bacterium causing Legionnaires’ Disease (LD) in humans. Rapid detection of Lp in water system is essential to reduce the risk of LD outbreaks. The methods currently available require expert skills and are time intensive, thus delaying intervention. In situ detection of Lp by biosensor would allow rapid implementation of control strategies. To this end, a biorecognition element is required. Aptamers are considered promising biorecognition molecules for biosensing. Aptamers are short oligonucleotide sequence folding into a specific structure and are able to bind to specific molecules. Currently, no aptamer and thus no aptamer-based technology exists for the detection of Lp. In this study, Systemic Evolution of Ligands through EXponential enrichment (SELEX) was used to identify aptamers binding specifically to Lp. Ten rounds of positive selection and two rounds of counter-selection against two Pseudomonas species were performed. Two aptamers binding strongly to Lp were identified with KD of 116 and 135 nM. Binding specificity of these two aptamers to Lp was confirmed by flow cytometry and fluorescence microscopy. Therefore, these two aptamers are promising biorecognition molecules for the detection of Lp in water systems.
Subject terms: Applied microbiology, Pathogens
Introduction
Legionella pneumophila (Lp) is a pathogenic Gram-negative bacterium responsible for two types of respiratory diseases, namely the severe pneumonia Legionnaires’ Disease (LD) and the milder flu-like Pontiac fever1. Lp occurs in both natural and engineered water systems and is one of the most prevalent pathogens in man-made, engineered water systems2. Infections occur when the bacteria are aerosolized, and the contaminated aerosols are inhaled, at which point Lp can then infect and replicate inside alveolar macrophages3. Modern water systems provide optimal transmission conditions for Lp by generating aerosols4. Leading sources of infection are cooling towers, hot water distribution systems, humidifiers, misters, showers, fountains, spa pools and evaporative condensers5.
Outbreaks of LD occur consistently globally and have increased in recent years. The average incidence rate is about 10–15 cases per million people6. According to the Centre for Disease Control, incidences of legionellosis have increased by four and a half times between 2000 and 20167. The Public Health Agency of Canada reports a 485% increase in the rate per 100,000 Legionellosis cases between the years 2000 to 20178. The rise in LD outbreaks can be attributed to several factors such as aging infrastructures and an aging population who is more vulnerable to such infections, as well as an increase in diagnosis and reporting of LD4,9. Most LD outbreaks, however, are the result of mismanagement of man-made water systems10. Examples of mismanagement of water distribution systems include keeping the temperature of the water below 50 °C and allowing water to stagnate10. In the case of cooling towers, a lack of regular cleaning and disinfection is associated with an increased risk of Lp spread10. In both cases, routine monitoring of Lp is critical to evaluate risk, initiate treatment of water systems, and prevent outbreaks10. The European Center of Disease Control (ECDC) specifies that immediate corrective measures must be taken when Lp levels reach a value of 10,000 CFU/L11.
Currently, there are two ISO-certified strategies to detect Lp from water systems: the standard plate count method (AFNOR NF T90-431, ISO 11731) and qPCR (AFNOR NF T90-471, ISO/TS 12869). The plate count method is the gold standard for detecting Lp and involves its cultivation on selective media and the enumeration of bacterial colonies showing Lp-specific morphology12–14. The whole procedure takes up to 14 days which delays the application of corrective measures and increases the chances of an outbreak15. A pilot study performed in 2011 evaluated the consistency of the results obtained by this method between several different laboratories. Qualitative results did not differ drastically between laboratories, but quantitative results showed large variation, within and between laboratories16. Therefore, the culture method should be used with caution to precisely enumerate Lp. A second major limitation is the presence of viable but non-culturable (VBNC) Lp cells which leads to an underestimation of the true amount of infectious Lp in a system17,18. The qPCR method relies on the quantification of Legionella DNA. Its major advantages in comparison with conventional culture method is the rapid turn-around time, high sensitivity and specificity, low limit of detection, as well as the ability to detect VBNC cells. When used in conjunction with the culture method, qPCR can serve as a powerful tool. There are, however, several drawbacks: qPCR typically overestimates Lp burden because it detects dead cells and the presence of PCR inhibitors may limit the use of this method19,20. In addition, multiple processing steps are required which increases the overall cost of the qPCR method21. Unfortunately, it is impossible to develop these two methods into rapid, cost-effective, sensitive tests that would detect Lp in real-time, on-site, without any additional processing steps22,23.
Biosensors are attractive detection technology that could address the problems associated with culture-based bacterial detection methods. These analytical devices are commonly used to assess and quantify in real-time, with high sensitivity, the presence of an analyte such as a protein, peptide or cell in a fluid24. However, a biosensing approach to Lp detection would require a specific biorecognition element, which, when coupled with a transducer, translates its interaction with Lp cells into a meaningful readout24.
Various biorecognition elements, such as antibodies, lectins or aptamers, can be used. The latter are becoming the primary choice for biosensing strategies due to their easily modifiable nature and versatility25,26. Aptamers are antibody analogues. They are short single stranded DNA or RNA oligonucleotides that can be cost effectively synthesized in a high throughput manner. The aptamer folds into a specific, stable structure and can interact with its targets via shape complementarity, hydrogen bonding, electrostatic interactions and stacking interactions27. This allows aptamers to bind with high affinity and specificity to a wide variety of targets ranging from small molecules, peptides, proteins to whole cells27. A key characteristic of aptamers is the possibility to generate them in vitro in the same condition as those used for detecting the analyte. This is a clear advantage over antibodies which are produced under strict physiological conditions28. In addition, aptamers can be easily modified and therefore be optimized for various sensing platforms such as lateral flow assays, surface plasmon resonance sensors, flow cytometry or fluorescence microscopy29.
The procedure by which an aptamer is created is known as Systemic Evolution of Ligands through EXponential enrichment (SELEX). Developed in 1990 by the teams of Gold and Szostak30,31, SELEX is an iterative process which involves incubating a target with a large library of oligonucleotides, separating the target bound and unbound oligonucleotides and then amplifying the target bound sequences via PCR for the next round of selection. The selection rounds are repeated until the oligonucleotide pool is enriched with sequences that bind specifically and with high affinity to the target30. Over the years, many variations of the original SELEX methods were published. Among those, one is particularly useful for the present study. Cell-SELEX can be used to select aptamers binding to whole living cells and, thus, eliminate the need for prior knowledge of a target molecule32,33. Rounds of counter-selection are typically used to reduce aptamer cross-reactivity across targets by eliminating non-specific aptamers29. Cell-SELEX has been successfully employed to isolate aptamers against various bacterial species such as E. coli, Salmonella typhimurium, Campylobacter jejuni, Listeria monocytogenes, Staphylococcus aureus and Vibrio parahaemolyticus28,33–38. Several of these aptamers are used in conjunction with optical, mechanical or electrical/electrochemical biosensors to mitigate the problems associated with traditional bacterial detection methods. Although numerous works have been done to detect Lp with the use of biosensors39–41 no aptamers binding to Lp have been reported yet. Consequently, no aptamer and thus no aptamer-based technology currently exists for the detection of Lp.
In this work, the cell-SELEX procedure was employed to generate aptamers binding to Lp. Two Pseudomonas species were used for counter-selection to improve the specificity of the aptamers. These species were chosen because they are γ-proteobacteria like Lp and are routinely found in premise plumbing and water systems where Lp is prevalent42,43. Two aptamers were identified and their binding affinity and specificity for Lp were evaluated by flow cytometry and fluorescence microscopy.
Materials and Methods
Bacterial strains and culture conditions
The environmental Lp strain lp120292, isolated from a cooling tower implicated in the 2012 outbreak in Quebec City, was used as the target strain for aptamer generation44. The strain Lp*GFP is lp120292 transformed with plasmid pXDC31 expressing the green fluorescent protein (GFP) under the Ptac promoter45. The thymidine auxotroph Lp strain Lp02, derived from Lp Philadelphia-1 was used to confirm binding of the aptamers46. Lp was cultured on CYE (ACES-buffered charcoal yeast extract) agar plates supplemented with 0.25 mg/ml L-cysteine and 0.4 mg/ml ferric pyrophosphate, at 37 °C for 3 days. Lp*GFP strain was grown on CYE media supplemented with 5 µg/ml chloramphenicol and 1 mM isopropyl β-D-1-thiogalactopyranoside. For liquid culture, Lp was suspended in AYE (ACES-buffered yeast extract) broth supplemented with 0.25 mg/ml L-cysteine and 0.4 mg/ml ferric pyrophosphate until post-exponential phase (OD600 of 2.5). Pseudomonas putida KT2440 and Pseudomonas fluorescens LMG1794, were first cultured on LB agar plates at 30 °C for 24 hours and then grown in LB medium (Difco) until the cultures reached post-exponential phase (OD600 of 2.0). Pseudomonas sp., Brevundiomonas sp., Bacillus sp., Staphylococcus sp. Sphingomonas sp., Stenotrophomonas sp. and Cupriavidus sp. were isolated from cooling towers as part of a different study (Paranjape, in preparation). Briefly, the bacteria were isolated on nutrient agar incubated at 30 °C for 24 h. Isolates were then classified using 16 S rDNA sequencing. These strains were first cultured on nutrient agar plates (Difco) at 30 °C for 24 hours and then grown in nutrient broth medium (Difco) overnight until the cultures reached post-exponential phase (OD600 of 2.0–2.5).
Oligonucleotides library and primers
A random ssDNA library of 1015 sequences was chemically synthesized and purified by HPLC (Integrated DNA Technologies). The library consisted of a central random region of 45 nucleotides flanked by two different primer binding regions at the 5′ and 3′ ends: 5′-GCAATGGTACGGTACTTCC-45N-CAAAAGTGCACGCTACTTTGCTAA-3′. The forward and reverse primers were conjugated with fluorescein (FITC) and biotin, respectively. The forward primer (FP) sequence is 5′-fluorescein-GCAATGGTACGGTACTTCC-3′. The reverse primer (RP) sequence is 5′-biotin-TTAGCAAAGTAGCGTGCACTTTTG-3′28. FITC was used to quantify ssDNA and monitor the SELEX procedure via flow cytometry. The biotin was used in conjunction with streptavidin coated magnetic beads (Promega) to generate ssDNA from the amplified double-stranded aptamer pool following PCR. For cloning and sequencing, PCR was performed with unmodified versions of the primers.
Bacterial cell-SELEX procedure
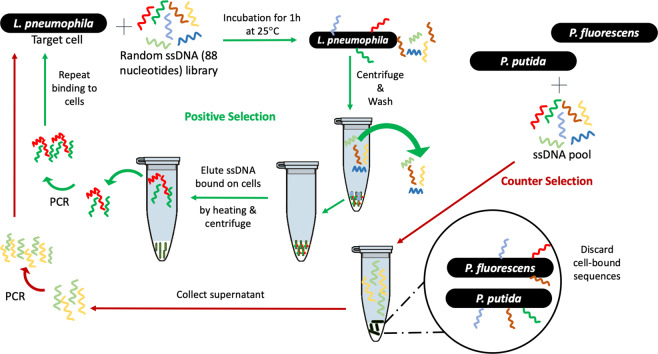
Cell-SELEX was performed as previously described28. Each round of SELEX consisted of three steps: Binding and elution, amplification, and recovery of ssDNA. Ten rounds of positive selection and two rounds of counter selection were performed (Fig. 1). The first round of counter selection was performed with P. putida KT2440; the second one with P. fluorescens LMG1794.
Figure 1.
Schematic illustration of bacterial cell-SELEX procedure used in this study. A random library of oligonucleotides is incubated with Lp lp120292 at room temperature for 1 h. Sequences that do not bind are washed off and the cell-bound sequences are then released and amplified via PCR. The resulting sequences are then submitted to another round of positive selection. P. fluorescens and P. putida were used to perform counter-selection rounds to eliminate non Lp-specific sequences.
Binding and Elution
Cell-SELEX was performed with cells suspended in an artificial freshwater medium (Fraquil) to replicate the physiological state of nutrient-limited environmental conditions47,48. Fraquil was prepared as described previously49 with a final iron concentration of 10 nM and filter-sterilized using a 0.2 µm filter (Sarstedt). Post-exponential phase cultures were rinsed twice with Fraquil (6000 g, 15 minutes) and suspended in Fraquil at an OD600 of 1 corresponding to a concentration of 109 CFU/ml. The concentration of cells was confirmed by CFU counts for each round. The suspension was incubated at room temperature for 24 h. Fraquil exposed cells were washed three times in 1X binding buffer (phosphate buffered saline with 0.1 mg/ml salmon sperm DNA, 1% bovine serum albumin, and 0.05% Tween 20) at room temperature (25 °C) using 6,000 g for ten minutes. The cell pellets were then suspended in 330 µl of 1X binding buffer. The aptamer pool was denatured by heating at 95 °C for 10 minutes, cooled immediately on ice for 10 minutes, and added to the cell suspension. Finally, 1X binding buffer was added to a total volume of 1 ml. For the first round, 32 µg of the initial library was used. For the subsequent rounds, approximately 400 ng of aptamer pool was used. The final mixture was incubated at 25 °C for 1 hour with mild shaking using a tube rotator at 150 rpm. Following incubation, the mixture was centrifuged at 6000 g for 10 minutes and washed twice with wash buffer (phosphate buffered saline containing 0.05% Tween 20) to remove unbound sequences. To elute the bound sequences from the cells, the final cell pellet was resuspended in 100 µl nuclease free water (Ambion) and heated at 95 °C for 10 minutes and immediately placed on ice for 10 minutes. After centrifuging at 6,000 g for 10 minutes at 25 °C, the supernatant was collected and purified using overnight ethanol precipitation at −20 °C with 5 µg of glycogen as a carrier to recover the eluted ssDNA. The pellet was recovered, dried and suspended in nuclease free water (Ambion). The concentration and quality of the ssDNA was determined using a Nanodrop spectrophotometer (Thermofisher). For counter-selection the supernatant containing the unbound sequences was collected and purified via ethanol precipitation, as described above. To ensure there was no amplification or collection of unwanted bacterial DNA (instead of the desired amplification and collection of ssDNA oligonucleotides), a control sample consisting of bacterial cells without aptamer was included in each round.
PCR amplification
The purified aptamer pool was then amplified by PCR with One Taq DNA polymerase (NEB), according to the manufacturer’s protocol. All primers were used at a final concentration of 0.5 µM. PCR conditions were as follows: initial heat activation at 95 °C for 5 min and 25 cycles of 95 °C for 30 s, 56.3 °C for 30 s, 72 °C for 10 s, and a final extension step of 10 min at 72 °C. After amplification, the concentration and size of the PCR product were confirmed by gel electrophoresis using a 2.0% agarose gel. PCR products were then purified using a MinElute PCR Purification Kit (Qiagen). As expected, no amplification was observed for the control samples, lacking aptamer template.
Recovery of ssDNA
Streptavidin coated magnetic beads (Promega Technology) were used, according to the manufacturer’s recommendation. Briefly, 600 µg of magnetic beads were washed twice and then resuspended in 900 µl of washing buffer (phosphate buffered saline with 0.05% Tween 20). Next, approximately1 µg of PCR product was incubated with the magnetic beads for 10 min, mixing gently by inversion after every few minutes. The mixture was then washed in 1 ml of washing buffer. Finally, the beads were incubated with 500 µl of 200 mM NaOH for 5 minutes. The supernatant was then collected, and the FITC-labelled ssDNA was purified using ethanol precipitation as mentioned previously and quantified with a Nanodrop spectrophotometer (Thermofisher).
Monitoring of SELEX by flow cytometry
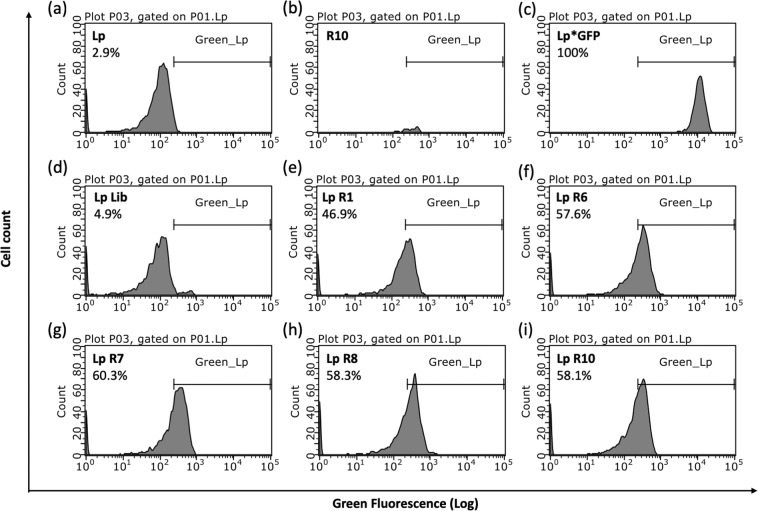
The binding of the FITC-labelled aptamer pools from rounds 1 (R1), 6 (R6), 7 (R7), 8 (R8) and 10 (R10) to Lp was assessed using flow cytometry. Briefly, 35 nM of aptamer pools from each of these rounds was incubated with 106 CFU/ml of Lp cells at 25 °C for 1 hour. Analysis was performed on a Guava easyCyte (Millipore) using the green fluorescence channel. A total of 5,000 events were recorded. Unlabeled cells were used as a control to measure autofluorescence. The Lp*GFP strain, producing strong green fluorescence from GFP, was used to adjust the gain of the green fluorescence channel. For analysis, a gate was first defined based on the forward and side scatters that included most of the cells. Then, a histogram of the number of cells vs the fluorescence intensity was used to define a region named Green_Lp where cells were considered positive for green fluorescence and therefore stained with aptamers. This region was setup to include very few cells of the unstained control and therefore represent fluorescence above the autofluorescence. Aptamer pools from R10 alone, without cells, was also analyzed to ensure that the aptamer alone was not forming aggregates that would be confused with cells.
Cloning and sequencing
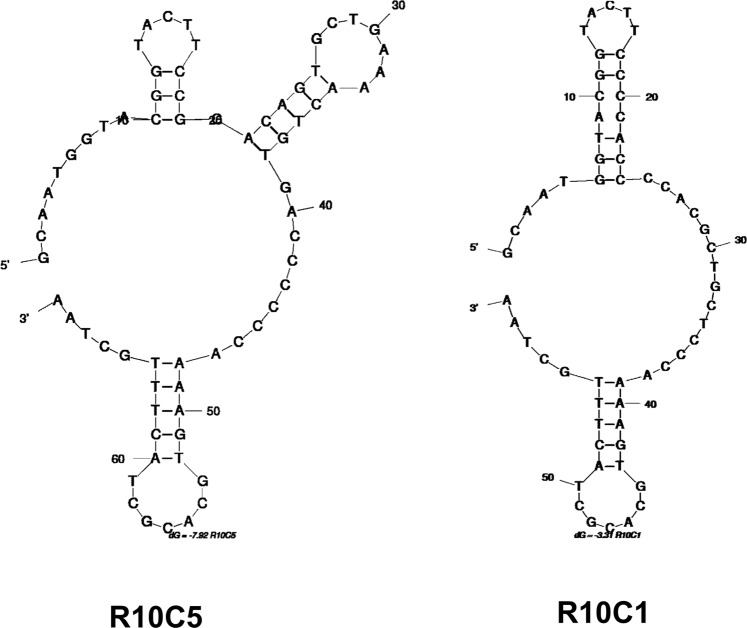
To identify sequences binding to Lp, the aptamer pool from the 10th round of SELEX was cloned with the pGEMT-easy Cloning and Ligation Kit (Promega). To investigate the effect of counter-selection on the aptamer pools, we also cloned and sequenced aptamers from the 6th round of SELEX. Positive colonies, containing aptamer inserts, were determined via blue-white screening and confirmed by PCR. Plasmids were extracted and purified using a Miniprep Kit (Qiagen) and sequenced by Sanger Sequencing at the Plate-forme d’Analyse Génomique of Laval University. Secondary structures of the aptamer sequences were determined using the Mfold web server using default parameters50.
Characterization of aptamers R10C5 and R10C1
The binding of the aptamers R10C5 and R10C1 to Lp and to the species used for counter-selection was further characterized. R10C5 and R10C1 were individually synthesized with FITC at the 5′ end (Integrated DNA Technology).
Determination of the disassociation constant (KD)
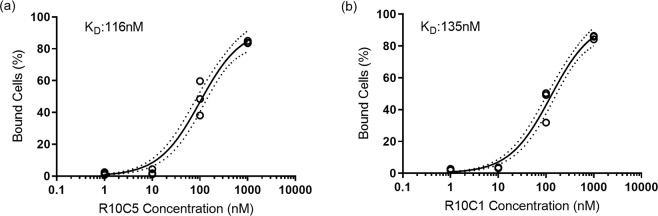
To determine the KD of R10C5 and R10C1, varying concentrations of FITC-tagged aptamers (1000 nM, 100 nM, 10 nM, and 1 nM) were incubated with 106 CFU/ml of Lp cells suspended in Fraquil and the fluorescence obtained at each concentration was measured using flow cytometry, as described above, in triplicate. The number of bound cells (FITC-positive) were recorded and used to determine the KD by interpolating the logarithmic curve using GraphPad Prism 7.03.
Specificity assay
To determine the specificity of R10C5 and R10C1 for Lp cells, the binding to counter-SELEX Pseudomonas strains as well as cooling tower isolates was tested using flow cytometry. All cells were suspended in Fraquil and prepared as described above for cell-SELEX. Briefly, 100 nM of R10C5 and R10C1 was incubated with 107 CFU/ml of the strain used for SELEX (lp120292), another Lp strain (Lp02), the strains used for counter-selection (P. putida KT2440 and P. fluorescens LMG1794), and the isolates from cooling towers (Pseudomonas sp., Brevundimonas sp., Bacillus sp., Staphlyococcus sp., Sphingomonas sp., Stenotrophomonas sp. and Cupriavidus sp.) for 1 hour at 25 °C with mild shaking. lp120292 was also incubated with 100 nM of a FITC-labeled scrambled sequence of aptamer R10C5 (5′-fluorescein-ACAGAATCAGTTCGAGTACATACGCGCGAAGACTCCTAAGGCCGTAGCGTTCTTCCCGGTAATACCATG) and R10C1(5′-fluorescein-TGTACTCCCGCGTCCCACCTGCTACCCGAAATAGAGTTTCCCTAGAAAGGCTTGCCCAAC). The suspension was centrifuged for 10 minutes at 6000 g to eliminate excess aptamer and resuspended in Fraquil. These suspensions were then analyzed using flow cytometry as described above. This experiment was done in triplicate. Cells suspended in Fraquil without any aptamer added were used as controls. The percentage of bound cells was determined as described above. Statistical differences were assessed using a one-way ANOVA and Dunnett correction for multiple comparison using GraphPad Prism 7.03.
Confocal fluorescence microscopy assay
FITC-labelled R10C5 and R10C1 aptamer (100 nM) were incubated with 108 CFU/ml of target cell lp120292 or counter-selection strain P. fluorescens LMG1794 for 1 hour at 25 °C on a tube rotator at 150 rpm. Cells were suspended in Fraquil as mentioned previously. Negative controls included cells suspended in Fraquil without any aptamers. The suspensions (10 µl) were dropped on a glass slide (Fisherbrand), and a #1.5 cover slip (VWR) was used to make a thin layer. The slides with suspensions were then transferred onto a microscope chamber and imaged using a Zeiss LSM 710 confocal microscope (Carl Zeiss, Oberkochen, Germany), a 100 X oil objective (Plan-Apochromat 100×/1.40 Oil DIC M27a) and a 488 nm argon laser (25 mW,). A pre-set FITC filter was used with excitation and emission wavelengths of 488 and 564 nm respectively. Images were analyzed using Fiji51.
Results and Discussion
Selection of aptamers binding to Lp
Cell-SELEX was used to select aptamers binding specifically to lp120292. This strain was selected because it was involved in the Quebec City Outbreak in 201244. To mimic the physiological state of Lp in a water system, Lp cells were grown to post-exponential phase and suspended in Fraquil for 24 h at 25 °C to induce starvation and the associated morphological and physiological changes47,48. Seven rounds of positive selection were performed, followed by one round of counter-selection, two rounds of positive selection, an additional round of counter-selection and a final round of positive selection (Fig. 1). Two Pseudomonas strains were used for counter-selection because they are also Gram-negative γ-proteobacteria frequently isolated from water systems where Lp is found as mentioned previously.
To monitor the progress of the SELEX procedure and ensure that the proportion of sequences binding to Lp was increasing, the binding of the FITC-labelled aptamer pools from rounds 1 (R1), 6 (R6), 7 (R7), 8 (R8) and 10 (R10) to Lp was examined using flow cytometry. Cells incubated with the initial aptamer library showed minimal fluorescence compared to the negative controls (Fig. 2d, Lp Lib). Cells incubated with aptamer from the first positive selection round showed a drastic increase in fluorescence (Fig. 2e, Lp R1). The saturation in the fluorescence intensity and the percentage of bound cells starting at R6 suggests that the pool is dominated by sequences binding to Lp (Fig. 2f–i, R6, R7, R8, R10). A small decrease in fluorescence and percentage of bound cells at R8 suggests that the first counter-selection step removed a few sequences. The fluorescence intensity remains similar between round 8 and 10 indicating that the second round of counter-selection did not remove Lp-specific sequences and that our strategy was successful in retrieving aptamers binding to Lp.
Figure 2.
Fluorescent labeling of Lp with FITC-labeled aptamers pools obtained after selected round of SELEX. Lp strain lp120292 was incubated without aptamers (Lp, a), with 35 nM of the aptamer library (Lp-lib, d) and with the aptamer pools obtained after round 1 (Lp-R1, e), 6 (Lp-R6, f), 7 (Lp-R7, g), 8 (Lp-R8, h) and round 10 (Lp-R10, i). The fluorescence obtained with aptamer from round 10 alone (R10, b), without cells, was also evaluated. Lp*GFP is a GFP producing version of lp120292 and is used as a positive control (c). The percentages refer to the proportion of cells with fluorescence above the autofluorescence, falling in the Green_Lp region.
Cloning and sequencing
Analyzing the sequences obtained from the 10th round of positive selection allowed for identifying two different ssDNA aptamers, named R10C5 and R10C1 (Table 1 and Fig. 3). Of the 13 sequences that were retrieved, 12 of them were R10C5 whereas 1 was R10C1. In contrast, the survey of a non-exhaustive list of the sequences present in the R6 aptamer pool revealed eight different sequences out of 9 clones, but none similar to R10C1 and R10C5. This illustrates the directional evolution of the pool as a result of the additional positive selection rounds and counter-selection steps52,53. A strong bottleneck effect was likely caused by the last four positive selection rounds and the apparently stringent counter-selection rounds, which most likely led to the removal of several aptamers.
Table 1.
Aptamer sequences from round 6 and round 10.
| Aptamer ID | Sequence | Frequency (%) |
|---|---|---|
| Before counter-SELEX (Round 6) | ||
| R6C1 | GCAATGGTACGGTACTTCCCCACTAACGCGCCCACGCACCCCTCGGCTACATCCAGCACCCGCCCAAAAGTGCACGCTACTTTGCTAA | 2/9 (22.2%) |
| R6C3 | GCAATGGTACGGTACTTCCCCACTCCACGCATCACAGCCTTTCACTGCCCACGCCTCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| R6C7 | GCAATGGTACGGTACTTCCACCACCGGAGTGTGCTTCAGCCGTGGTACAATACTGCCGTGTATCCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| R6C11 | GCAATGGTACGGTACTTCCCCCACTGCACACACAAAGGGCCAGCATCAACACACGCGCCGTTCCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| R6C12 | GCAATGGTACGGTACTTCCCACCCCGCCACGCCGATAGCCTCCCATACTCCCCCCGCANGTCCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| R6C15 | GCAATGGTACGGTACTTCCCGCGCACCCCACACCTCCGCACACCGCATGCCTCCCCTTAGGCCCCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| R6C16 | GCAATGGTACGGTACTTCCCACTGCCGAACGCGCCCTCTCCTGCTGCCTCCACACATGGTCGCCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| R6C18 | GCAATGGTACGGTACTTCCCCCACCAAGCCCATACACGTACAGCCTACCACAATCCACATCGGGCCAAAAGTGCACGCTACTTTGCTAA | 1/9 (11.1%) |
| Post Counter-SELEX (Round 10) | ||
| R10C5 | GCAATGGTACGGTACTTCCGGACAGTGCTGAAAACTGTGACCCCCCAAAAGTGCACGCTACTTTGCTAA | 12/13 (92.3%) |
| R10C1 | GCAATGGTACGGTACTTCCCCACCCCACGCTGCTCCCAAAAGTGCACGCTACTTTGCTAA | 1/13 (7.7%) |
Figure 3.
The structure of the aptamers R10C5 and R10C1 were determined using Mfold.
Determination of KD
The calculated KD is 116 nM for R10C5 and 135 nM for R10C1 (Fig. 4). These values are comparable to high affinity antibodies that typically show nanomolar ranges of KD for small protein targets54. These values are also comparable to values of published aptamers created against whole bacterial pathogens. For example, aptamers isolated against Escherichia coli, Enterobacter aerogenes, Klebsiella pneumoniae, Citrobacter freundii, Bacillus subtilis, and Staphylococcus epidermidis showed KD ranging from 9.22–38.5 nM55. Additionally two 62 nuccd to Staphylococcus aureus with KD of 35 nM and 129 nM56.
Figure 4.
Determination of the KD of the aptamers R10C5 (a) and R10C1 (b). Lp (lp120292) was incubated with 10-fold dilutions of the FITC-tagged aptamers and the fluorescence was measured by flow cytometry. The number of cells displaying fluorescence above the autofluorescence were counted as bound cells, as described in Fig. 2. The graphs show individual values of three experiments. The equilibrium dissociation constant, KD was calculated using GraphPad Prism 7.03. The non-linear regression (solid line) with 95% confidence interval error envelopes (dotted lines) is shown.
Specificity of R10C5 and R10C1
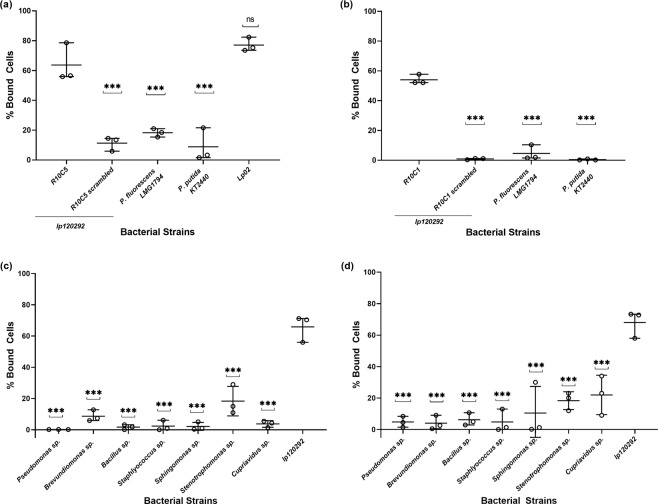
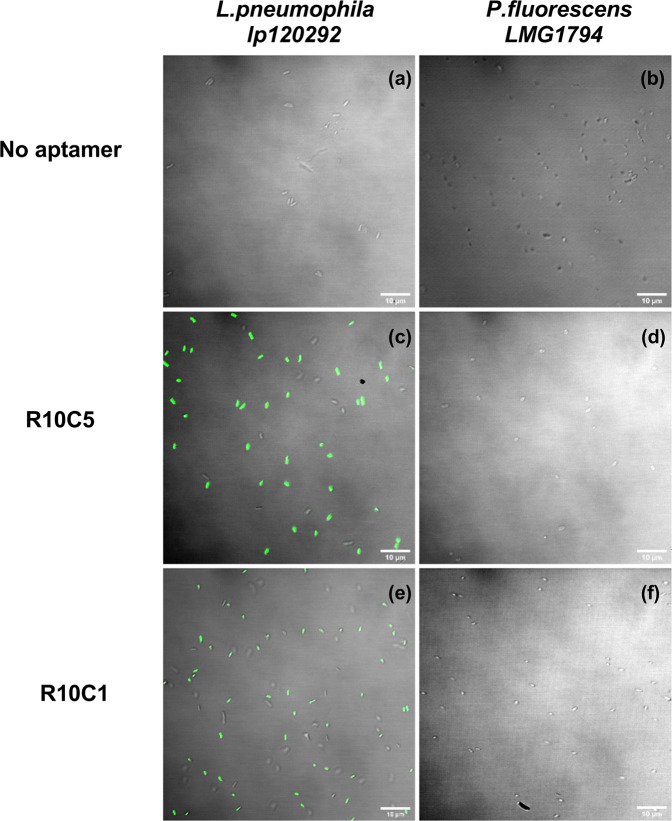
Figure 5a shows the binding of R10C5 and R10C1 to the strains used for counter-selection. Around 60% of lp120292 cells are stained by R10C5, consistent with previous results shown in Fig. 2, but only 20% of Pseudomonas strains are labelled. Similarly, R10C1 shows significantly more binding to Lp than to Pseudomonas (Fig. 5b). Of note, the scrambled sequences of aptamer R10C5 and R10C1 bind minimally to lp120292 (Fig. 5a,b). The aptamer R10C5 stained Lp02 similarly to lp120292, suggesting that the binding of this aptamer is not restricted to a particular strain of Lp (Fig. 5a). Moreover, both aptamers show very low binding to environmental isolates from cooling tower water. For the majority of the isolates, less than 10% of cells were labeled by the aptamers (Fig. 5c,d). The specificity of these aptamers for Lp was further analyzed by confocal fluorescence microscopy. Both aptamers strongly stained Lp (Fig. 6) but not P. fluorescens LMG1794, one of the strains used for counter-selection. These results support the notion that the aptamers R10C5 and R10C1 are highly specific to Lp.
Figure 5.
The specificity of FITC-labelled R10C5 and R10C1 aptamers binding to Lp strain lp120292 (positive control), to counter SELEX Pseudomonas strains (a,b) as well as to environmental isolates (c,d) was analyzed by flow cytometry. The binding of R10C5 to Lp strain Lp02 was also analyzed. The percentage of cells bound by R10C5 (a,c) and R10C1 (b,d) to counter-SELEX strains (a,b) and environmental isolates (c,d) are presented. The binding of a scrambled sequence of aptamer R10C5 and a scrambled sequence of aptamer R10C1 to Lp strain lp120292 was also investigated (scrambled). The values of three experiments are shown with the mean and standard deviation. A one-way ANOVA with a Dunnett correction for multiple comparisons was used to infer statistical significance compared to Lp strain lp120292: ***P < 0.001; ns, not significant.
Figure 6.
The specificity of FITC-labelled R10C5 and R10C1 aptamer was tested by measuring their ability to bind to Lp strains lp120292 (a,c and e) and to P. fluorescens LMG1794 (b,d and f) by confocal fluorescence microscopy. The No aptamer controls consists of cells alone (a,b). Cells were incubated with aptamer R10C5 (c,d) and aptamer R10C1 (e,f).
In conclusion, our cell-SELEX strategy was successful in identifying two aptamers binding to Lp with high affinity (KD = 116 nM for R10C5 and 135 nM for R10C1). Whereas R10C5 seems to stain Lp more strongly then R10C1, the latter seems more specific to Lp, showing minimal binding to the counter SELEX Pseudomonas strain. Both aptamers showed minimal binding to cooling towers isolates, indicating that the aptamers are suitable to detect Lp in complex water samples. Modification of these aptamers could be attempted to further increase their affinity and specificity to Lp. Based on the results presented here, these aptamers are promising candidates as biorecognition elements to develop a biosensor to detect Lp in real time and in situ.
Acknowledgements
We would like to acknowledge the McGill Institute of Parasitology for use of the Zeiss LSM 710 confocal microscope. We would also like to thank Jonathan Perreault (INRS-IAF) for useful discussion and Youssef Chebli (McGill University) for all his extensive assistance with the microscope. The Pseudomonas strains used for counter selection are a kind gift from Eric Déziel (INRS-IAF). This study was supported by an NSERC Strategic Partnership Grant to MT and SPF. MS was supported by a CRIPA scholarship supported by the Fonds de recherche du Québec - Nature et technologies n°RS-170946.
Author contributions
M.S., M.T. and S.P.F. designed the study. M.S. and S.P.F. planned the experiments. M.S. and D.C. performed the experiments. M.S. wrote the first draft of the manuscript. M.S., M.T. and S.P.F. edited the manuscript. All authors approved the submitted version of the manuscript.
Competing interests
All the work described in this manuscript is the subject of patent applications filed in USA, patent application no US 16/850,355; and in Canada – patent application number pending at the time of revised manuscript submission. The inventors are Mariam Saad, Maryam Tabrizian and Sebastien P. Faucher. At the time of submission of the manuscript, the applications were under review. The authors declare no other competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McDade JE, et al. Legionnaires Disease. New England Journal of Medicine. 1977;297(22):1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 2.Kozak, N. A. et al. “Identification of Legionella in the Environment”. Methods in Molecular Biology. Humana Press, 3-25 (2013). [DOI] [PubMed]
- 3.Albert-Weissenberger C, et al. Legionella pneumophila a human pathogen that co-evolved with fresh water protozoa. Cellular and Molecular Life Sciences. 2007;64(4):432–448. doi: 10.1007/s00018-006-6391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parr A, et al. Legionellosis on the Rise. Journal of Public Health Management and Practice. 2015;21(5):E17–E26.. doi: 10.1097/PHH.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Heijnsbergen E, et al. Confirmed and Potential Sources of Legionella Reviewed. Environmental Science &. Technology. 2015;49(8):4797–4815. doi: 10.1021/acs.est.5b00142. [DOI] [PubMed] [Google Scholar]
- 6.MacIntyre CR, et al. Rolling epidemic of Legionnaires’ disease outbreaks in small geographic areas. Emerging Microbes & Infections. 2018;7(1):1–10. doi: 10.1038/s41426-018-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Center for Immunization and Respiratory Diseases, Division of Bacterial Diseases. ‘Legionella: History Burden and Trends’, Center for Disease Control. https://www.cdc.gov/legionella/about/history.html (2018).
- 8.Canadian Notifiable Diseases Surveillance System (CNDSS), Public Health Agency of Canada (PHAC). ‘Rate per 100,000 of reported cases over time in Canada, Legionellosis’, Accessed 12.2.2020 https://diseases.canada.ca/notifiable/charts?c=pl (2020).
- 9.Diederen BM. Legionella spp. and Legionnaires disease. Journal of Infection. 2008;56(1):1–12. doi: 10.1016/j.jinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds Kelly, Sexton Jonathan. Legionellosis Prevention and Response Training for Environmental Health Professionals. American Journal of Infection Control. 2017;45(6):S60–S61. [Google Scholar]
- 11.European Guidelines Working Group (EGWG). “European technical guidelines for the prevention, Control and Investigation of Infections caused by Legionella species. 2017.” In. (2019).
- 12.Bopp CA, et al. Isolation of Legionella spp. from environmental water samples by low-pH treatment and use of a selective medium. Journal of Clinical Microbiology. 1981;13(4):714–719. doi: 10.1128/jcm.13.4.714-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CDC, Centers for Disease Control and Prevention. ‘Procedures for the recovery of Legionella from the environment’, Division of Bacterial and Mycotic Diseases RDLS. Atlanta. (2005)
- 14.ISO. 2017. ‘Water quality–Enumeration of Legionella’, ISO 11731 (2017)
- 15.Trudel, L. et al. Management of the 2012 Legionella crisis in Quebec City: need for a better communication between resources and knowledge transfer. Frontiers in Microbiology5 (2014). [DOI] [PMC free article] [PubMed]
- 16.Lucas CE, et al. Accuracy and precision of Legionella isolation by US laboratories in the ELITE program pilot study. Water Research. 2011;45((15):4428–4436. doi: 10.1016/j.watres.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Epalle T, et al. Viable but Not Culturable Forms of Legionella pneumophila Generated After Heat Shock Treatment Are Infectious for Macrophage-Like and Alveolar Epithelial Cells After Resuscitation on Acanthamoeba polyphaga. Microbial Ecology. 2015;69(1):215–224. doi: 10.1007/s00248-014-0470-x. [DOI] [PubMed] [Google Scholar]
- 18.Li, L. et al. The importance of the viable but non-culturable state in human bacterial pathogens. Frontiers in Microbiology5 (2014). [DOI] [PMC free article] [PubMed]
- 19.Diaz-Flores Á, et al. Comparing methods of determining Legionella spp. in complex water matrices. BMC Microbiology. 2015;15:1. doi: 10.1186/s12866-015-0423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor M, et al. Limitations of Using Propidium Monoazide with qPCR to Discriminate between Live and Dead Legionella in Biofilm Samples. Microbiology Insights. 2014;7:MBI.S17723. doi: 10.4137/MBI.S17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiley H, et al. Legionella detection by culture and qPCR: Comparing apples and oranges. Critical Reviews in Microbiology. 2016;42(1):65–74. doi: 10.3109/1040841X.2014.885930. [DOI] [PubMed] [Google Scholar]
- 22.Deshmukh RA, et al. Recent developments in detection and enumeration of waterborne bacteria: a retrospective minireview. MicrobiologyOpen. 2016;5((6):901–922. doi: 10.1002/mbo3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law, J. W. F. et al. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Frontiers in Microbiology5 (2015). [DOI] [PMC free article] [PubMed]
- 24.Ahmed A, et al. Biosensors for Whole-Cell Bacterial Detection. Clinical Microbiology Reviews. 2014;27(3):631–646. doi: 10.1128/CMR.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davydova A, et al. Aptamers against pathogenic microorganisms. Critical Reviews in Microbiology. 2015;42(6):847–865. doi: 10.3109/1040841X.2015.1070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, et al. Whole-cell based aptamer selection for selective capture of microorganisms using microfluidic devices. Analytical Methods. 2015;7((15):6339–6345. [Google Scholar]
- 27.Gelinas AD, et al. Embracing proteins: structural themes in aptamer protein complexes. Current Opinion in Structural Biology. 2016;36:122–132. doi: 10.1016/j.sbi.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Kim YS, et al. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cellsystematic evolution of ligands by exponential enrichment approach. Analytical Biochemistry. 2013;436(1):22–28. doi: 10.1016/j.ab.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Rozenblum GT, et al. Aptamers: current challenges and future prospects. Expert Opinion on Drug Discovery. 2016;11.2:127–135. doi: 10.1517/17460441.2016.1126244. [DOI] [PubMed] [Google Scholar]
- 30.Ellington AD, et al. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 31.Tuerk C, et al. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 32.Darmostuk M, et al. Current approaches in SELEX: An update to aptamer selection technology. Biotechnology Advances. 2015;33(6):1141–1161. doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Kaur H, et al. Recent developments in cell-SELEX technology for aptamer selection. Biochimica et Biophysica Acta (BBA) - General Subjects. 2018;1862(10):2323–2329. doi: 10.1016/j.bbagen.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 34.Ahn JY, et al. Surface Plasmon Resonance Aptamer Biosensor for Discriminating Pathogenic Bacteria Vibrio parahaemolyticus. Journal of Nanoscience and Nanotechnology. 2018;18(3):1599–1605. doi: 10.1166/jnn.2018.14212. [DOI] [PubMed] [Google Scholar]
- 35.Duan N, et al. Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella Typhimurium and Staphylococcus aureus. Analytica Chimica Acta. 2012;723:1–6. doi: 10.1016/j.aca.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Duan N, et al. Selection, identification and application of a DNA aptamer against Listeria monocytogenes. Food Control. 2013;33(1):239–243. [Google Scholar]
- 37.Manzano M, et al. OLED-based DNA biochip for Campylobacter spp. detection in poultry meat samples. Biosensors and Bioelectronics. 2015;66:271–276. doi: 10.1016/j.bios.2014.11.042. [DOI] [PubMed] [Google Scholar]
- 38.Moon J, et al. Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers. Sensors. 2015;15(4):8884–8897. doi: 10.3390/s150408884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enrico DL, et al. SPR based immunosensor for detection of Legionella pneumophila in water samples. Optics Communications. 2013;294:420–426. [Google Scholar]
- 40.Melaine F, et al. Selective and High Dynamic Range Assay Format for Multiplex Detection of Pathogenic Pseudomonas aeruginosa, Salmonella typhimurium, and Legionella pneumophila RNAs Using Surface Plasmon Resonance Imaging. Analytical Chemistry. 2017;89(14):7802–7807. doi: 10.1021/acs.analchem.7b01942. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi, N. et al. Rapid on-site monitoring of Legionella pneumophila in cooling tower water using a portable microfluidic system. Scientific Reports7(1) (2017). [DOI] [PMC free article] [PubMed]
- 42.Khweek, A. et al. Factors Mediating Environmental Biofilm Formation by Legionella pneumophila. Frontiers in Cellular and Infection Microbiology8 (2018). [DOI] [PMC free article] [PubMed]
- 43.Stewart CR, et al. Legionella pneumophila Persists within Biofilms Formed by Klebsiella pneumoniae, Flavobacterium sp., and Pseudomonas fluorescens under Dynamic Flow Conditions. Plos One. 2012;7(11):e50560. doi: 10.1371/journal.pone.0050560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lévesque S, et al. Molecular Typing of Legionella pneumophila Isolates in the Province of Quebec from 2005 to 2015. Plos one. 2016;11(10):e0163818. doi: 10.1371/journal.pone.0163818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hovel-Miner G, et al. σ S Controls Multiple Pathways Associated with Intracellular Multiplication of Legionella pneumophila. Journal of Bacteriology. 2009;191(8):2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berger KH, et al. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Molecular Microbiology. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 47.Li L, et al. Transcriptomic changes of Legionella pneumophila in water. BMC Genomics. 2015;16(1):637. doi: 10.1186/s12864-015-1869-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendis N, et al. Short-Term and Long-Term Survival and Virulence of Legionella pneumophila in the Defined Freshwater Medium Fraquil. Plos One. 2015;10(9):e0139277. doi: 10.1371/journal.pone.0139277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morel, F. et al. “Description of the algal growth media ‘Aquil’and ‘Fraquil’. Water Quality Laboratory, Ralph Parsons Laboratory for Water Resources and Hydrodynamics, Massachusetts Institute of Technology.” Technical Report 16 (1975).
- 50.Zuker M, et al. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31(13):3406-3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schütze T, et al. Probing the SELEX Process with Next-Generation Sequencing. Plos one. 2011;6(12)):e29604. doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoltenburg R, et al. Refining the Results of a Classical SELEX Experiment by Expanding the Sequence Data Set of an Aptamer Pool Selected for Protein A. International Journal of Molecular Sciences. 2018;19(2):642. doi: 10.3390/ijms19020642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, et al. Aptamers as targeted therapeutics: current potential and challenges. Nature Reviews Drug Discovery. 2017;16(3):181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song MS, et al. Detecting and Discriminating Shigella sonnei Using an Aptamer-Based Fluorescent Biosensor Platform. Molecules. 2017;22(5):825. doi: 10.3390/molecules22050825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang YC, et al. Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles. Scientific Reports. 2013;3:1. doi: 10.1038/srep01863. [DOI] [PMC free article] [PubMed] [Google Scholar]