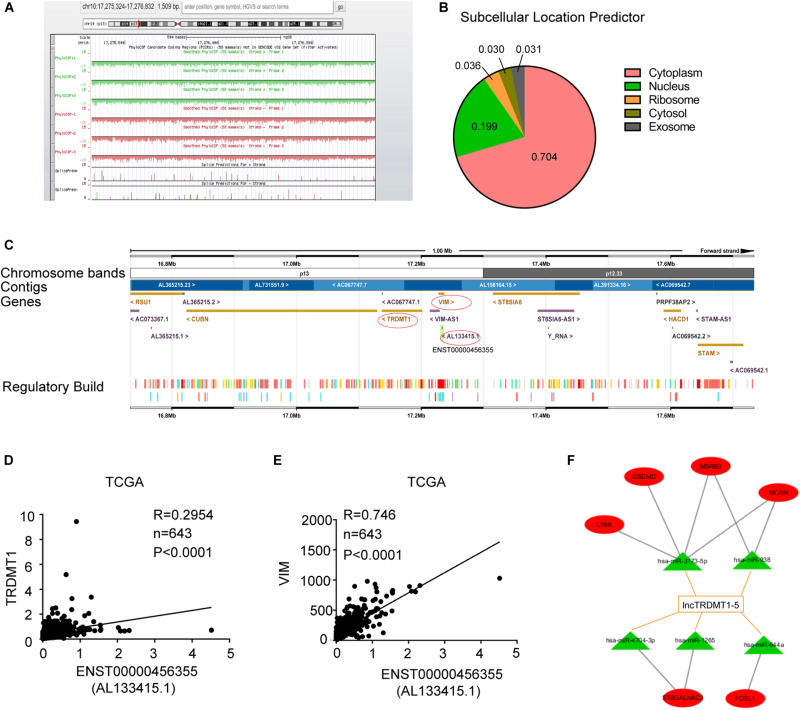
FIGURE 6.
Subcellular location and functional ceRNA network prediction for lnc-TRDMT1-5. (A) Schematic annotation of the lnc-TRDMT1-5 genomic locus on chromosome 10q17,275,324-17,276,832 in humans. Coding potency was analyzed using PhyloCSF. Scores above zero suggest coding potential, whereas scores below zero represent non-coding potential. (B) The pie chart shows the subcellular location scores of lnc-TRDMT1-5 using a subcellular location predictor (lncLocator) (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) according to the sequence analysis. (C) The region of ENST0000456355 (AL133415.1) was identified in detail by the Ensembl database (http://asia.ensembl.org), and it shows that the protein coding gene VIM shared part of the location site with AL133415.1. Another coding gene, TRDMT1, was upstream of AL133415.1. (D) A correlation analysis between lnc-TRDMT1-5 and TRDMT1 in the TCGA database was performed on 590 invasive BC and 53 normal breast tissues. (E) The correlation analysis between lnc-TRDMT1-5 and VIM was performed on 590 invasive BC and 53 normal breast tissues. (F) A correlation network comprising lnc-TRDMT1-5-miRNA-mRNA. Correlation between lncTRDMT1-5 and miRNAs were screened for the criteria of correlation scores >0.9, and binding scores >0.04. Correlations between selected miRNAs and mRNAs were calculated and identified by using the TargetScan database. A total of five miRNAs and six mRNAs were selected with connecting edges using Cytoscape 3.6.