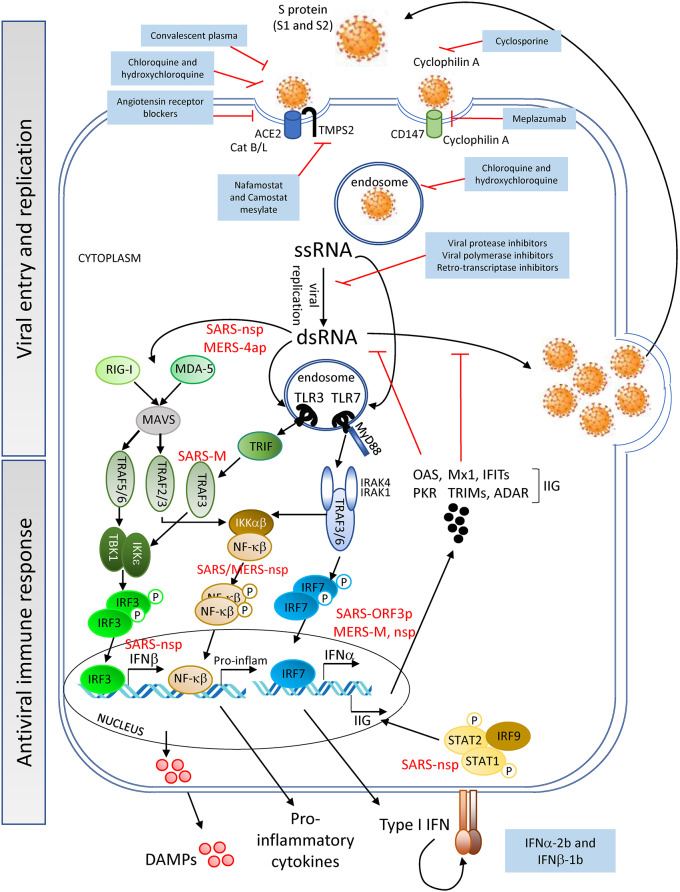
Figure 1.
Viral entry and replication, and host antiviral immune response—SARS-CoV-2 (Spike (S)-protein) recognizes cell surface proteins (ACE2, TMPS2, and CD147) which facilitate the endocytosis of the virus particle. Viral genome (ssRNA) exits the endosomal vesicles and starts the replication cycle generating dsRNA intermediates, protein translation, encapsidation and generation of new viral particles. New virions can subsequently infect neighboring cells. DAMPs are being released by the dying cells into the extracellular space. Innate immune receptors such as TLR3, RIG-I and MDA-5 or TLR7 can sense viral RNA (dsRNA or ssRNA, respectively) and initiate a signaling cascade for the production of IFNα/β and pro-inflammatory cytokines. Interferon-induced genes (IIG) will be expressed as a result of this type I IFN feedback loop, blocking viral replication. SARS-CoV and MERS-CoV structural and non-structural proteins can interfere with the cells' innate immune response [see proteins in red; for further information refer also to the Kindler et al. (29)], delaying the production of sufficient levels of antiviral cytokines to prevent control of viral replication at the early stages. SARS-CoV-2 might utilize similar evasion mechanisms, although no data have yet been reported. Pharmacological interventions (blue-background boxes) are being proposed to block both the entry and genomic replication of SARS-CoV-2, as well as to boost the innate immune response.