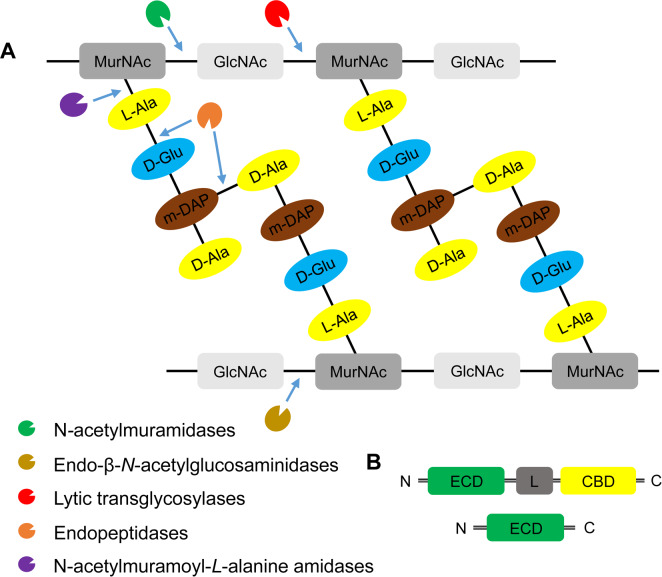
Fig. 3.
The domain structures, diversity, and the mode-of-action of endolysins. (A) Peptidoglycan structure and targets of five classes of endolysins. Peptidoglycan is a heteropolymer of alternating amino sugars N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MucNAc), and tetrapeptide side chains, L-alanine (L-Ala), D-glutamate (D-Glu), meso-diaminopimeric acid (m-DAP) or L-lysine, and D-alanine (D-Ala), attached to the lactyl group of muramic acid. Depending on the enzymatic specificity, endolysins can be divided into five classes, N-acetylmuramindase (lysozymes or muramidases), endo-β-N-acetylglucosaminidase (glucosaminidases), N-acetylmuramoyl-L-alanine amidase (NAM-amidases), endopeptidases and lytic transglycosylases. Bonds cleaved by different endolysins were indicated by blue arrows. (B) Domain structures of bacteriophage endolysins. Ones derived from bacteriophages infecting Grampositive-bacteria include an enzymatic catalytic domain (ECD) and at least one cell-wall binding domain (CBD) and connected by a flexible linker (L) (Nelson et al., 2012). By contrast, endolysins from bacteriophages infecting Gram-negative bacteria are mostly globular structure with single ECD without a specific CBD (Pohane and Jain, 2015). N, N-terminus; C, C-terminus.