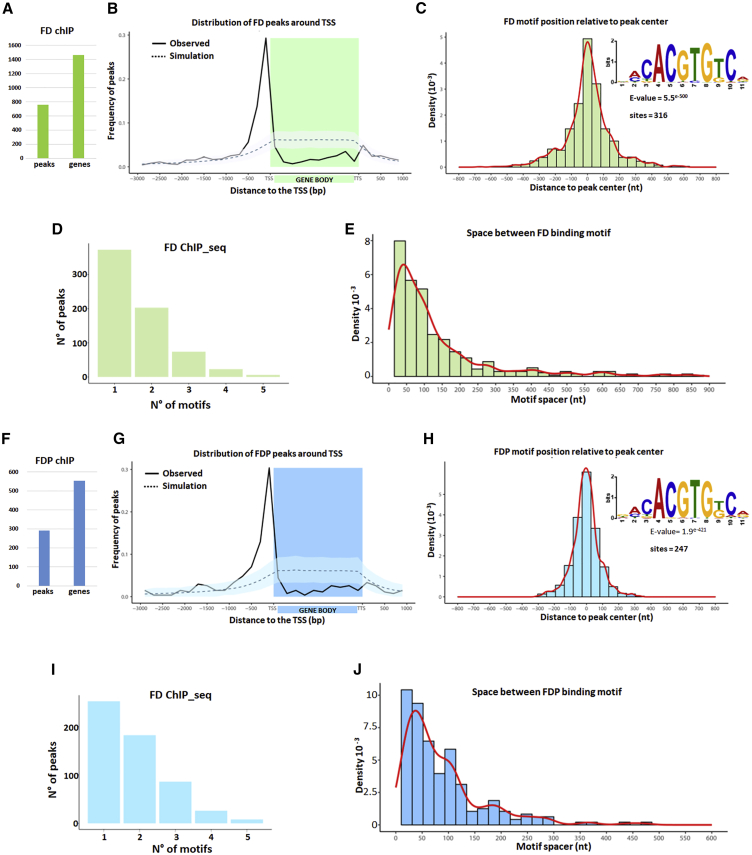
Figure 5.
Genome-wide Target Sites of FD and FDP
(A–E) FD genome-wide targets. (A) The number of FD ChIP-seq peaks and associated genes. (B) Distribution of FD peaks in a region between 3 kb upstream of the transcription start site (TSS) and 1 kb downstream of the transcription termination site (TTS) of the closest gene. The solid line shows the positional distribution of observed FD peaks. The observed positional distribution was compared to those of 1,000 randomly generated peak sets. As the distributions were determined using bins, both the mean (dashed line) and the 95% confidence interval (light shaded color) for each bin are depicted. (C) Density plot of distances between the center of the G-boxes and the center of FDP peaks. The red line marks the shape of the distribution. The inset shows the logo of the enriched sequence motif identified by MEME motif analysis. The E-value indicates the statistical significance of the identified motif. (D) Number of FD ChIP-seq peaks containing the indicated number of G-box motifs. (E) Distance between neighboring G-box motifs in FD targets. The red line marks the shape of the distribution.
(F–J) FDP genome-wide targets. (F) Predicted number of FDP peaks and corresponding associated genes. (G) Distribution of FDP peaks as described for FD in (B). (H) Density plot of distances of G-boxes to the center of FDP peaks, as described for FD in (C). (I) Number of FD ChIP-seq peaks containing the indicated number of G-box motifs. (J) Distance between neighboring G-box motifs in FDP targets.