Figure 6.
Characterization of FD and FDP Target Genes
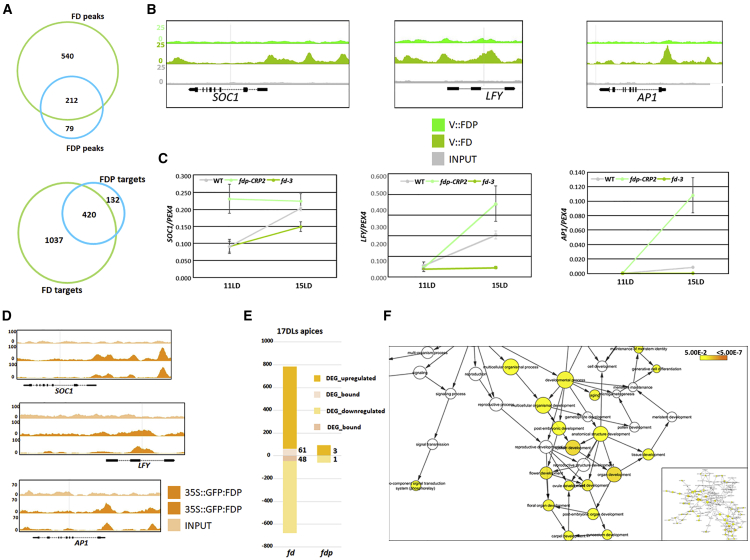
(A) Venn diagram illustrating common peaks (top panel) and genes (bottom panel) in the FD and FDP ChIP-seq datasets.
(B) FD and FDP binding profiles to flowering genes. The three panels display FD and FDP and the control (INPUT) peaks at SOC1, LFY, and AP1 visualized with the Integrated Genome Browser (IGB).
(C) qRT-PCR analysis of SOC1, LFY, and AP1 mRNA abundance in apices of fd-3 and fdp-CRP2 mutants in 11- and 15-day-old plants under LDs.
(D) FDP-binding profiles to flowering genes when expressed from the 35S promoter.
(E) Proportion of DEGs (adj. p ≤ 0.05) that are up- or downregulated in the RNA-seq of 17 LD apices of fd-3 and fdp-CRP2. The number of DEGs directly bound by FD or FDP is illustrated in digits.
(F) GO terms enriched within the 109 genes differentially expressed and bound by FD using apices of 17-day-old plants. The color of the circles reflects the p value and the size the overrepresentation or underrepresentation of GO categories according a hypergeometric test (false discovery rate [FDR] < 0.05). The inset on the left shows an overview of all GO terms.