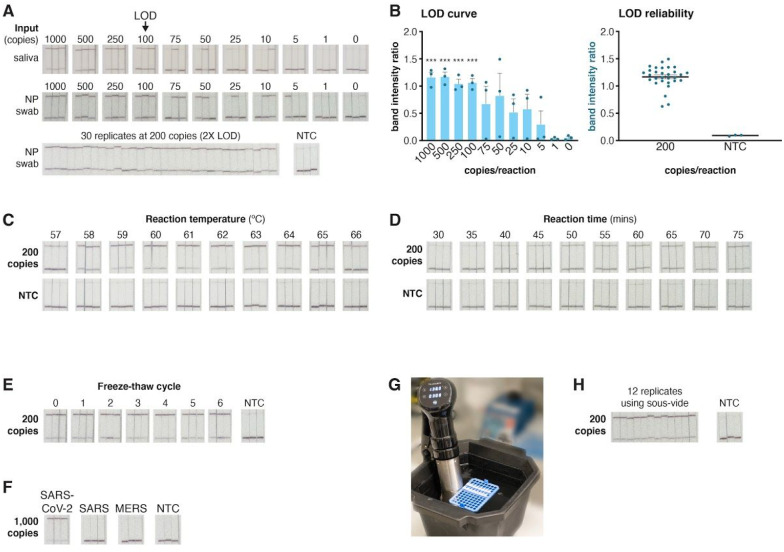
Figure 4: STOPCovid performance on lateral flow strips.
A. Determination of the limit of detection (LOD) for STOPCovid with lateral flow readout using three replicates per condition (top). Synthetic genomic standards spiked into saliva or nasopharyngeal (NP) swab was used for input. All three replicates could be positively detected (top band on strip) down to 100 SARS-CoV-2 standard genomes per reaction. At twice the determined LOD of 100 genomic copies per reaction in NP swab, STOPCovid yielded positive results for 30 replicates (bottom). NTC, no template control.
B. Quantification of the band intensity ratio (top band/bottom band) for the LOD determination reactions (left) and 30x replicates (right) from panel A. ***, p < 0.001.
C. Effect of reaction temperature on STOPCovid lateral flow detection for 200 SARS-CoV-2 copies per reaction.
D. Effect of reaction incubation time on STOPCovid lateral flow detection for 200 SARS-CoV-2 copies per reaction.
E. Effect of mastermix freeze-thaw cycles on STOPCovid lateral flow detection for 200 SARS-CoV-2 copies per reaction.
F. Evaluation of cross-reactivity for COVID-19 STOPCovid lateral flow test for SARS and MERS N genes. All inputs were at 1,000 copies per reaction.
G. Example STOPCovid setup using a 60°C water bath powered by a simple commercial sous-vide device.
H. STOPCovid lateral flow detection of SARS-CoV-2 at 200 copies per reaction using a 60°C sous-vide water bath.