Figure 3.
Molecular Phenotypes Discriminate Clinical from gnomAD Missense PTEN Variation
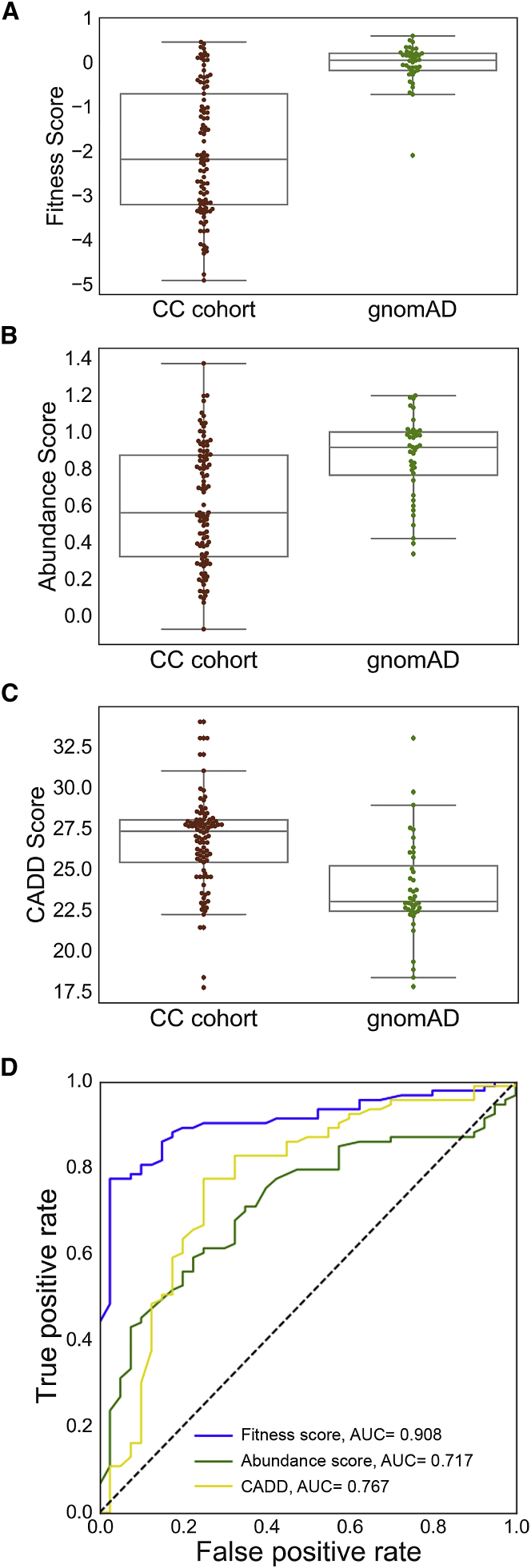
These analyses are done at the variant level so that recurrent variants do not bias the results. Two known pathogenic variants were removed from the gnomAD list (Materials and Methods).
(A)Boxplots comparing fitness scores of variants found in the CC cohort to the scores of variants found in gnomAD. Cohen’s r = 0.61.
(B) Boxplots comparing abundance scores of variants found in the CC cohort to the scores of variants found in gnomAD. Cohen’s r = 0.37.
(C) Boxplots comparing CADD scores of variants found in the CC cohort to the scores of variants found in gnomAD. Cohen’s r = 0.45.
(D) Receiver operator characteristic curves for univariate models. The feature weights are reported in Table S7.