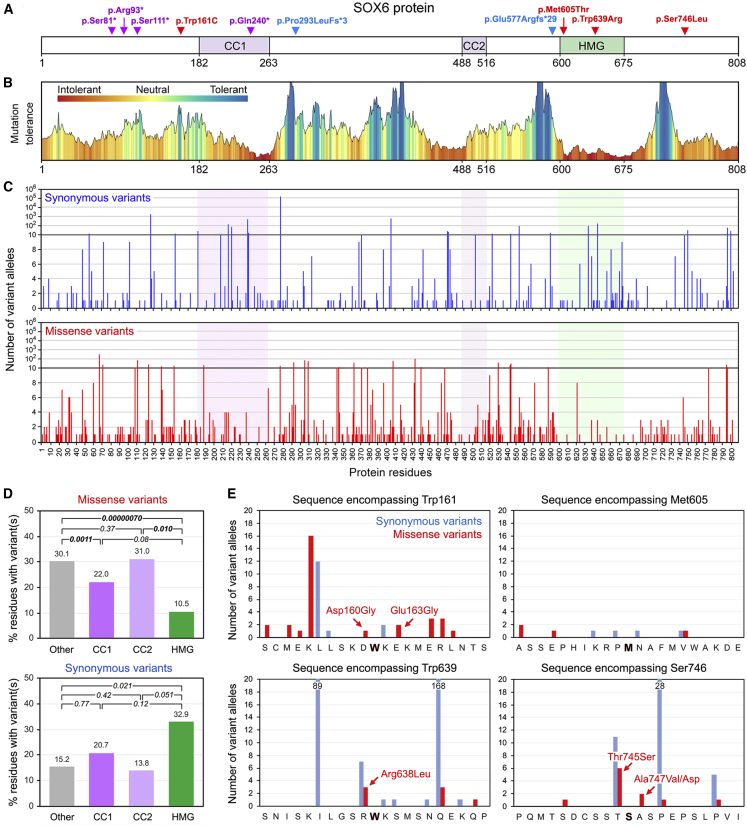
Figure 3.
Analysis of SOX6 SNVs in Affected Individuals 9–18 and in gnomAD Individuals
(A) Location of study subjects’ SNVs in the SOX6 isoform encoded by the GenBank: NM_033326.3 transcript. The protein and domain residue boundaries are indicated underneath the schematic. CC1, primary coiled-coil domain; CC2, secondary coiled-coil domain; HMG, HMG domain. Red represents missense variants, purple represents nonsense variants, and blue represents frameshift variants.
(B) Plot of the mutation tolerance of SOX6 residues downloaded from MetaDome.
(C) Counts and distribution of SOX6 synonymous and missense variants found in gnomAD individuals.
(D) Percentages of residues carrying at least one missense or synonymous variant in the functional and other domains of SOX6 in gnomAD individuals. We performed paired t tests to calculate the statistical significance of differences between domains. p values are indicated.
(E) Numbers of synonymous and missense variants detected in gnomAD individuals in the sequences encompassing the missense variants identified in four study subjects. The nature of the missense variants closest to the residues altered in the four affected individuals is indicated.