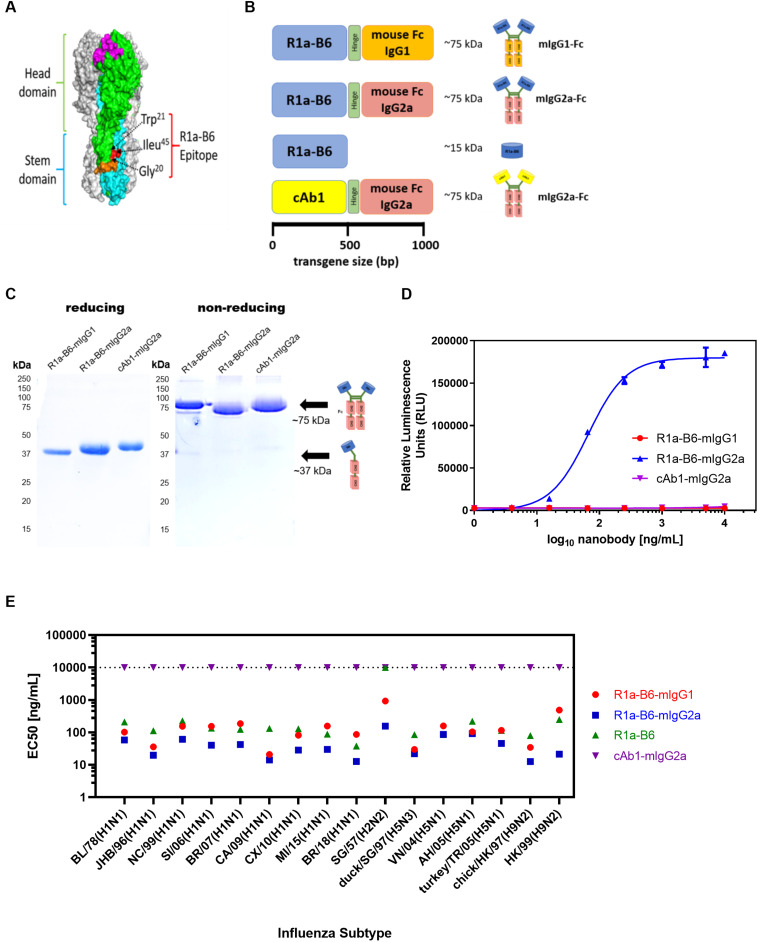
FIGURE 1.
R1a-B6 reformatted for in vivo gene delivery. (A) Surface structure model of hemagglutinin (HA) trimer of A(H1N1)pdm09 (PDB structure 3AL4) showing the key epitope residues of R1a-B6, Gly20, Trp21, and Ile45 (shown in red) located in the HA stem region (cyan) (40). The receptor binding site (magenta), fusion peptide (orange), and head domain (green) are also illustrated. (B) Four constructs, (i) R1a-B6 mouse Fc IgG1, (ii) R1a-B6 mouse Fc IgG2a, (iii) monovalent R1a-B6, and (iv) negative control mouse Fc IgG2a fusion carrying a nanobody, cAb1 [adapted from Arbabi Ghahroudi et al. (54)], specific for chicken egg white lysozyme were produced in vitro. These constructs were cloned into an AAV expression system for protein expression in vivo. (C) Expression and purification of nanobody-Fc fusions. Detection of proteins was carried out under reducing and non-reducing conditions in SDS-PAGE gels. Theoretical molecular weights (MW) for R1a-B6-mIgG1, R1a-B6-mIgG2a, and cAb1-mIgG2a are ∼37 kDa under denaturing (reducing) conditions, and ∼75 kDA under non-reducing conditions. (D) In vitro ADCC activation. Activation of luciferase reporter gene is shown in relative luminescence units (RLU) as a function of nanobody-Fc concentration. Each well was measured in triplicate. (E) Binding of R1a-B6 against a broad range of influenza A subtypes as tested by ELISA. Half maximal effective concentration (EC50) was measured in duplicate. There was no binding on A/TX/12(H3N2) or B/Brisbane/08 (data not shown). All values above the dotted line indicate no binding activity.