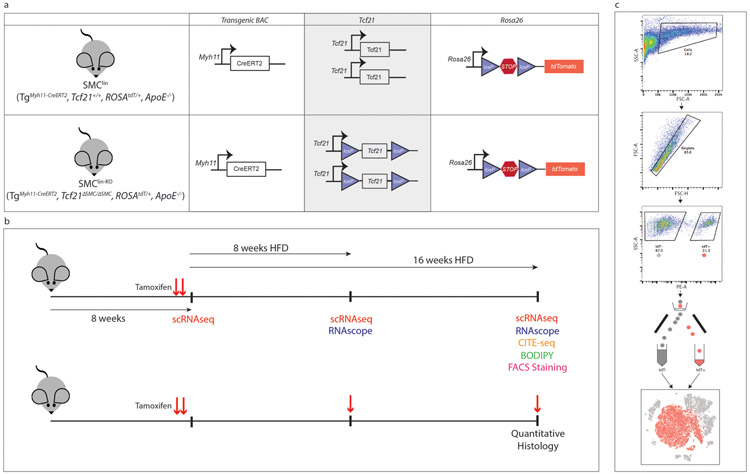
Extended Data Figure 1. Design of mouse experiments.
(a) Alleles present in SMClin and SMClin-KO mice. KO (knockout) refers to Tcf21, lin = lineage tracing, Tg = transgene, ΔSMC = SMC cell-specific KO (b) Mice were maintained on chow diet from birth until 7 weeks of age, then underwent gavage and high-fat diet (HFD) treatment. For single-cell RNAseq (scRNAseq), RNAscope, CITE-seq, histology involving BODIPY and FACS staining experiments (upper timeline), mice were gavaged only at 7 weeks of age, prior to onset of HFD, as denoted by red arrows. For scRNAseq experiments, mice were sacrificed at baseline (72 hours after initial tamoxifen gavage), or after 8 weeks or 16 weeks of HFD. For RNAscope experiments, mice were sacrificed after either 8 weeks or 16 weeks of HFD. For the CITE-seq experiment, mice were sacrificed after 16 weeks of HFD. For BODIPY studies, mice were sacrificed after 16 week of HFD. For the FACS staining experiment two mice, one after 12 weeks HFD and another after 15 weeks HFD were used. For quantitative histology experiments (lower timeline), mice were gavaged at 7 weeks of age, after 8 weeks of HFD and after 16 weeks of HFD (48 hours prior to sacrifice) as denoted by red arrows. For these quantitative histology experiments, all mice were sacrificed after 16 weeks of HFD. (c) Fluorescence activated cell sorting (FACS) workflow for isolating single cells from the mouse aortic root.