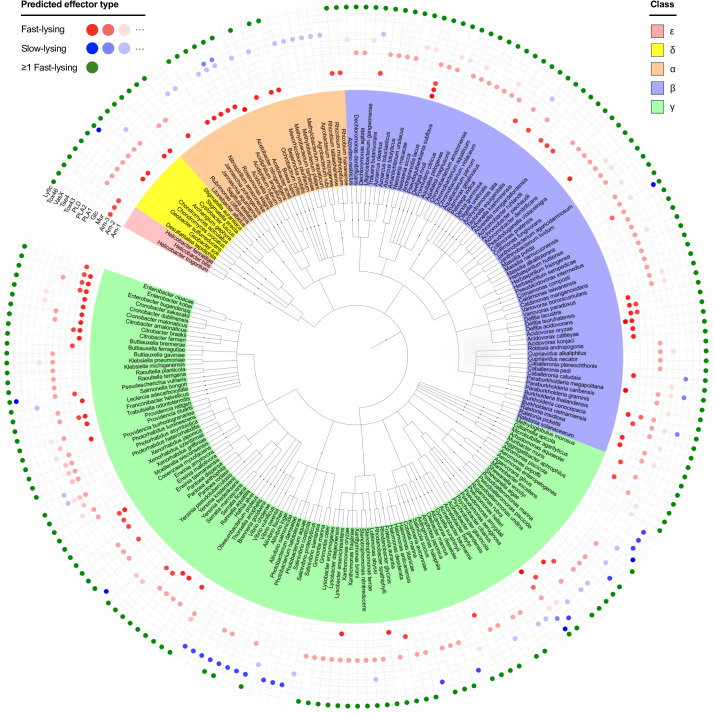
Fig 4. Taxonomy showing distribution of lytic and nonlytic T6SS toxins across the Proteobacteria.
Each species is positioned in the tree according to its taxonomic classification in the NCBI taxonomy database and colored according to Proteobacterial Class (α, β, δ, ε, γ). Colored circles indicate the T6SS effectors associated with a single, randomly chosen strain from each species. Effectors are labeled according to their molecular target (see below); effectors expected to induce rapid cell lysis (Am-1-3, Mur, Glc, PLA1-2, PLD; “fast-lysing”) are shaded in red; those expected to lyse cells more slowly (Tox43, Tse4, VasX, Tox46; “slow-lysing”) are shaded blue. A green marker signifies that a given strain has at least 1 fast-lysing effector in its repertoire. Each species in this dataset is represented by 1 strain only, and so these data are not representative of the full toxin repertoire of any given species. However, by comparing example strains across many species, one can robustly assess preference for known lytic toxins. Plotting data from all 474 species was not feasible in 1 figure, so we plot approximately half (222) of the strains here, retaining at least 1 representative from each genus. The full tree is available to download as a separate file (S1 Data). Original data are from LaCourse and colleagues [40]; reanalyzed data are available from dx.doi.org/10.6084/m9.figshare.11980491. Am-1-3, peptidoglycan amidases; Mur/Glc, peptidoglycan glucosidases; NCBI, National Center for Biotechnology Information; PLA1-2 and PLD, phospholipases; Tox43, DNases; Tox46; NAD(P)+ glycohydrolases; Tse4/VasX, pore-forming.