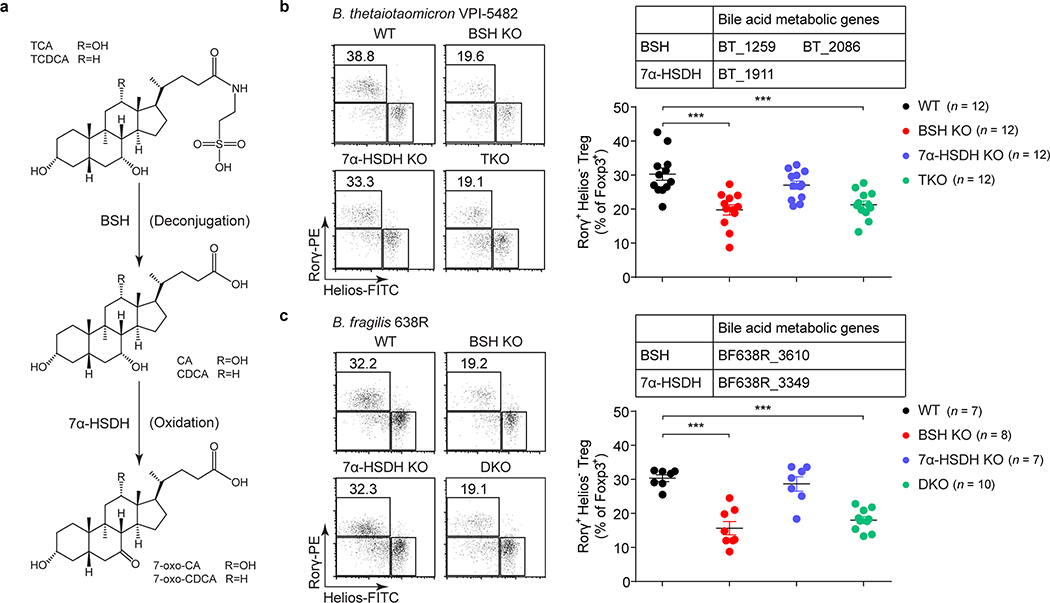
Fig. 2. Gut bacteria control colonic RORγ+ Tregs through their bile acid metabolic pathways.
(a) Schematic diagram of BA metabolic pathways in B. thetaiotaomicron and B. fragilis. (b) Each of 4 groups of GF mice was colonized with one of the following microbes for 2 weeks: 1) a wild-type (WT) strain of B. thetaiotaomicron; 2) a BSH mutant strain (BSH KO, in which both the BT_1259 and BT_2086 genes are deleted); 3) a 7α-HSDH mutant strain (7α-HSDH KO, in which the BT_1911 gene is deleted); or 4) a triple-mutant strain (TKO, in which all three genes are deleted). Representative plots and frequencies of RORγ+Helios– in the Foxp3+CD4+TCRβ+ Treg population are shown. (c) Each of 4 groups of GF mice was colonized with one of the following microbes for 2 weeks: 1) a WT strain of B. fragilis; 2) a BSH KO strain (in which the BF638R_3610 gene is deleted); 3) a 7α-HSDH KO strain (in which the BF638R_3349 gene is deleted); or 4) a double-mutant strain (DKO, in which both genes are deleted). Colonic Tregs were analyzed as in b. Data are pooled from three independent experiments in b, c. n represents biologically independent animals. Bars indicate mean ± SEM values in b, c. ∗∗∗p < 0.001 in one-way analysis of variance followed by the Bonferroni post hoc test.