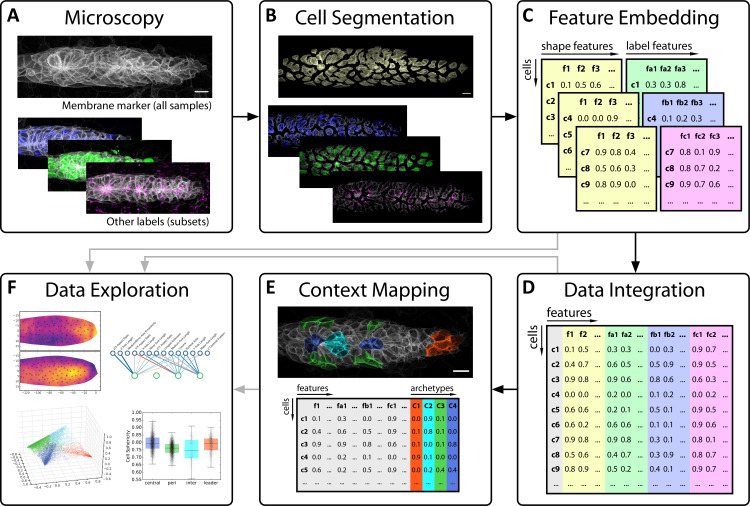
Figure 1. Overview of key steps in our data-driven analysis workflow.
(A) Image data of the tissue of interest are acquired using 3D confocal fluorescence microscopy. Each sample is labeled with a membrane marker to delineate cell boundaries (top) and samples can additionally be labeled with various other markers of interest (bottom, colored). (B) Using an automated image analysis pipeline, single cells are automatically segmented based on the membrane marker to prepare them for analysis, illustrated here by shifting them apart. (C) Next, data extraction takes place to arrive at numerical features representing the cell shapes (yellow) and the various fluorescent protein distributions of additional markers (other colors). (D) Such well-structured data simplify the application of machine learning techniques for data integration, which here is performed based on cell shape as a common reference measurement. (E) A similar strategy can be used to map manually annotated contextual knowledge (top) into the dataset (bottom), in this case specific cell archetypes chosen based on prior knowledge of the tissue's biology. (F) Finally, all of the resulting data are explored and interpreted through various visualizations and statistics.