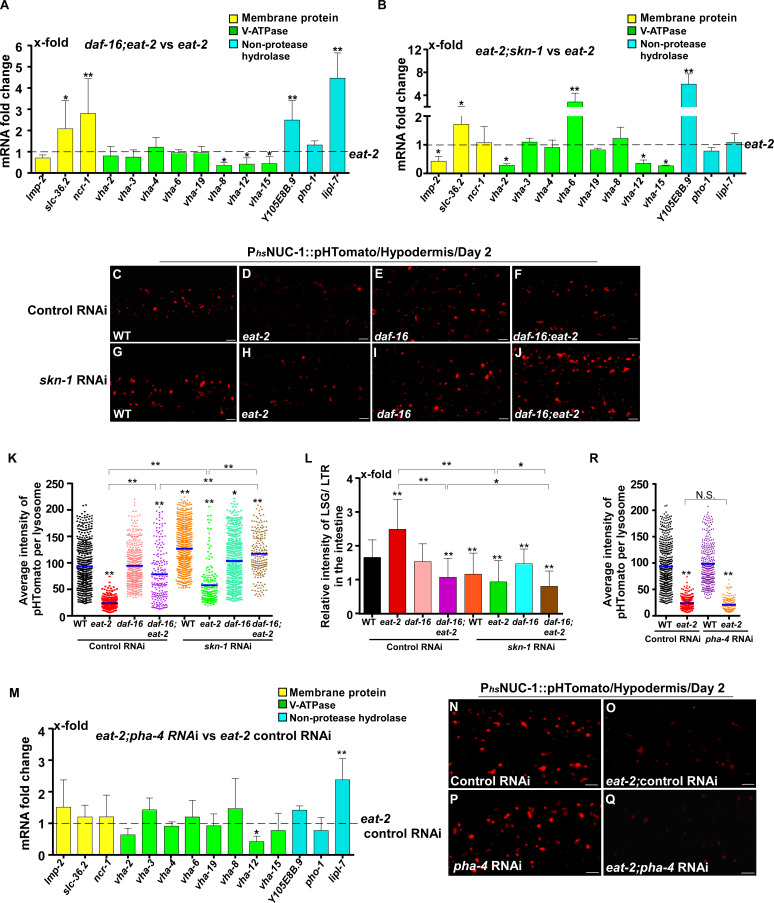
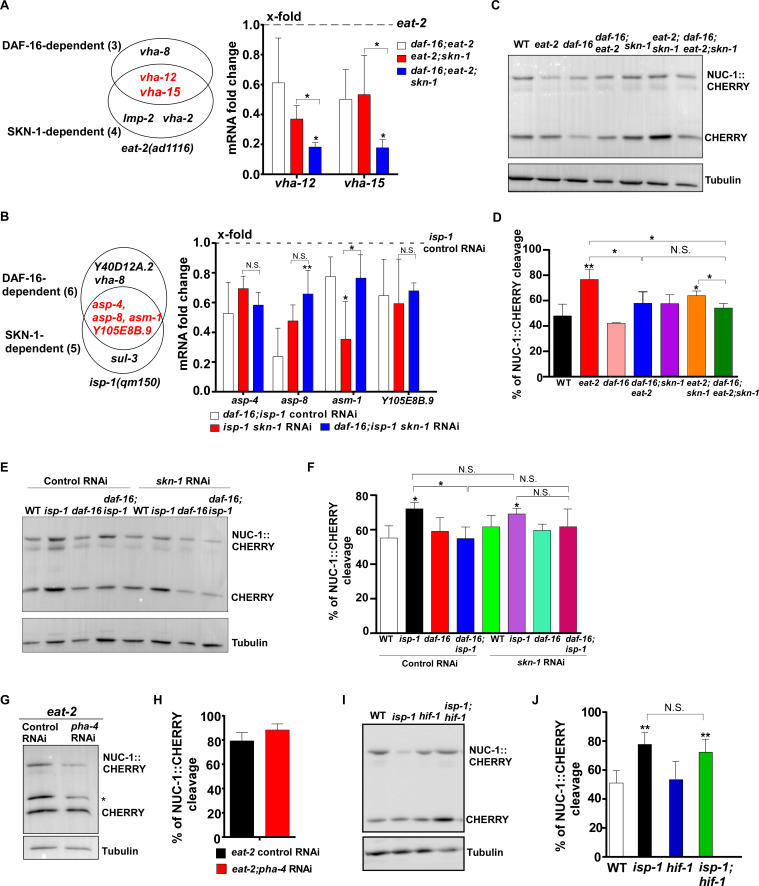
Figure 7. DAF-16 and SKN-1, but not PHA-4, regulate lysosomal acidity and gene expression in eat-2 mutants.
(A, B, M) Expression of the 14 upregulated lysosomal genes in eat-2(ad1116) was analyzed by qRT-PCR in daf-16;eat-2 (A), eat-2;skn-1 (B) and eat-2;pha-4 RNAi (M) worms at day 1. Three independent experiments were performed. The transcription level of lysosomal genes in eat-2(ad1116) (A, B) or eat-2(ad1116) control RNAi (M) at day 1 was normalized to ‘1’ for comparison. (C–J, N–Q) Confocal fluorescence images of the hypodermis in the indicated strains expressing NUC-1::pHTomato controlled by the heat-shock (hs) promoter. Scale bars: 5 μm. The average intensity of pHTomato per lysosome was quantified (K, R). At least 20 animals were scored in each strain. (L) The relative intensity of LSG/LTR in the intestine was quantified in the indicated strains at day 2. At least 10 animals were scored in each strain. In (A, B, K, L, M, R), data are shown as mean ± SD. Multiple t testing (A, B, M) or one-way ANOVA with Tukey's multiple comparisons test (K, L, R) was performed to compare datasets of double mutants with eat-2 (A, B), or eat-2 control RNAi (M), or to compare all other datasets with wild type treated with control RNAi (K, L, R), or datasets that are linked by lines (K, L, R). *p<0.05; **p<0.001. All other points had p>0.05. N.S., no significance.