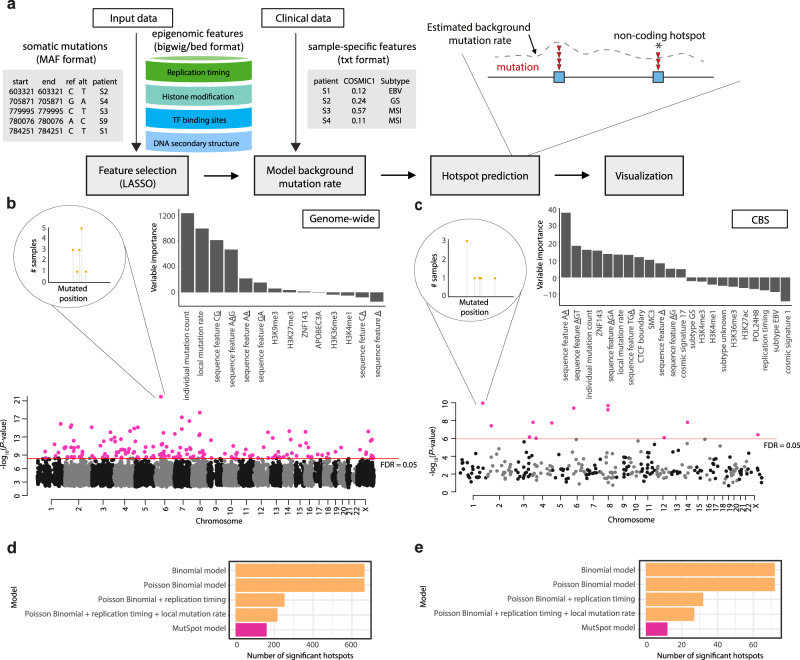
Fig. 1. MutSpot analysis on 168 gastric cancer whole genomes.
a MutSpot analysis workflow. b, c For each analysis, MutSpot outputs three types of descriptive figures: a Manhattan plot, a feature importance plot of features evaluated by the background mutation model, and lollipop plots of the top hotspots. Figures produced by MutSpot from b a genome-wide analysis and c a CBS-specific analysis of 168 gastric cancer whole genomes. Hotspots with FDR <0.05 are labeled in magenta. d, e Comparison of the number of hotspots detected using MutSpot with the number of hotspots detected using other statistical approaches in d the genome-wide and e CBS-specific analyses.