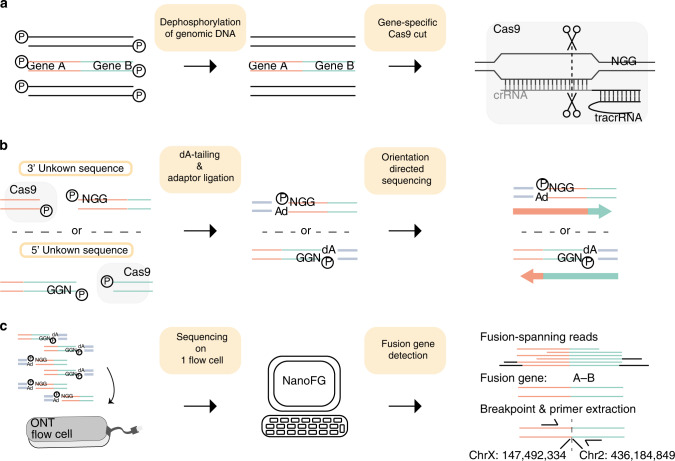
Fig. 1. Schematic overview of FUDGE.
a Genomic DNA sample is dephosphorylated and crRNA-guided target-specific double-stranded cuts are introduced through Cas9. b Phosphorylation-sites are exposed through the double-strand breaks; however, Cas9 remains bound to the PAM-distal side of the cut and blocks phosphorylation of the DNA on this side. DNA-ends are dA-tailed and adapters are ligated only to phosphorylated DNA-ends proximal to the PAM sequence. Sequencing direction is dictated by the adapters, towards the unknown sequence. c Targeted libraries are loaded and sequenced on one ONT flow cell (R9.4). NanoFG is run on the nanopore sequencing data, extracts fusion-spanning reads, detects fusion genes and provides exact fusion gene configuration, breakpoint-location, breakpoint-sequence and fusion-spanning primer sequences.