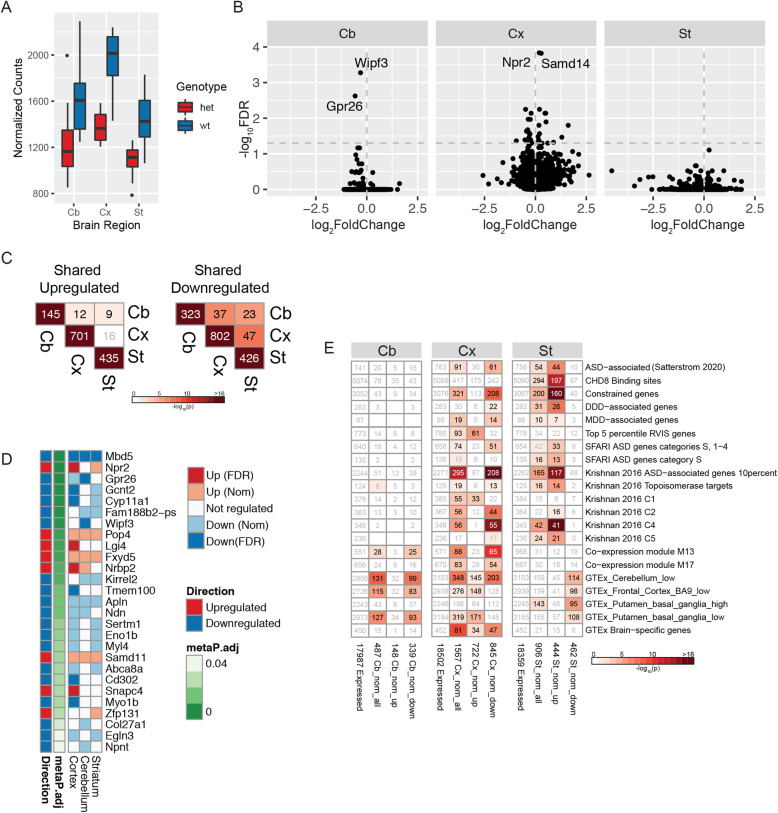
Fig. 2.
Differential expression analysis of RNA-seq data from mouse brain regions. a Expression of Mbd5 mRNA as normalized counts in three brain regions: cerebellum (Cb), striatum (St), and cortex (Cx). b Volcano plots of differential expression analysis per region. c Gene overlaps in differentially expressed genes in mouse brain regions. The color indicates the significance (−log10(p)) of Fisher’s enrichment test, and the number shows the number of overlapping DEGs at nominal significance. Only the genes that were present in all tests were considered as background for enrichment tests and for overlap analyses. d Meta-analysis of p values from differential gene expression comparisons in each brain region using Fisher’s method. The color in the heatmap shows whether the gene was significantly up- or downregulated in a corresponding brain region. “Direction” indicates whether the gene was analyzed in a group of upregulated genes or downregulated genes. “metaP.adj” shows the p value of meta-analysis using Fisher’s method. e Enrichments of differentially expressed genes at FDR and at nominal significance in gene sets previously associated with brain development and developmental disorders. The description of gene lists is provided in Supplementary Tables 5 & 6.