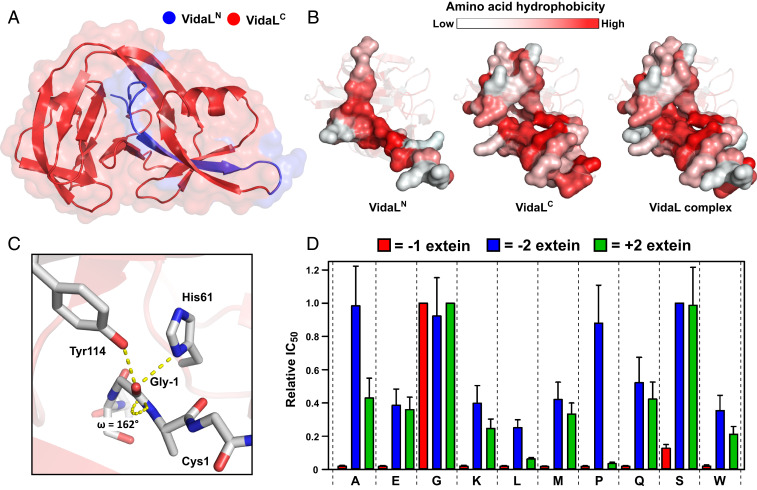
Fig. 2.
X-ray crystal structure of VidaL informs extein dependence. (A) A 1.65-Å–resolution crystal structure of an engineered fused version of VidaL bearing inactivating mutations. The regions corresponding to the N- and C-intein fragments in the split version are colored blue and red, respectively. (B) Space-filling models displaying the interaction between VidaLN and VidaLC with residues colored by hydrophobicity (23). The binding is largely driven by hydrophobic (red) interactions. (C) View of the N-extein of VidaL, displaying H-bonding interactions between Gly-1, His61, and Tyr114 resulting in −1 amide bond distortion (ω = 162°). (D) Extein dependence of VidaL PTS explored using an E. coli-based antibiotic selection assay. Histogram displaying the relative IC50 values of kanamycin for each of the extein mutants tested at the −1 (red), −2 (blue), and +2 (green) extein residues (mean ± SD, n = 3). The value for the wild-type residue at each position is normalized to 1.