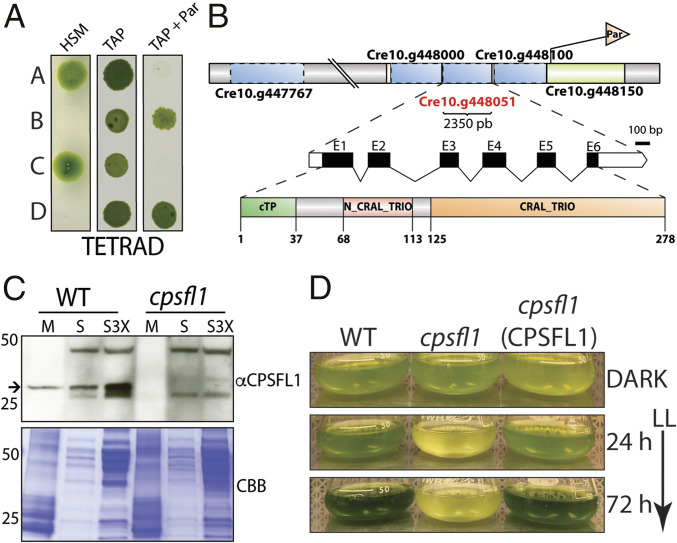
Fig. 1.
Genetic analysis and light sensitivity of the cpsfl1 mutant. (A) Analysis of a representative tetrad from a back-cross of the mutant (mating type +) to the WT (mating type −). Cells were spotted onto minimal (HSM) and acetate-containing (TAP) solid media, in the presence or absence of paromomycin, and grown under constant light (80-μmol photons m−2 s−1) and in the dark, respectively. (B) Schematic representations showing the paromomycin-resistance gene insertion site within chromosome 10 in the mutant, the complete annotation of the CPSFL1 gene, Cre10.g448051, as well as the C. reinhardtii CPSFL1 protein showing the N-terminal chloroplast transit peptide (cTP), the N-terminal N-CRAL-TRIO, and the C-terminal CRAL-TRIO protein domains. (C) Immunoblot analysis from WT and cpsfl1 fractionated cells showing the absence of the CPSFL1 protein in the mutant. The membrane fraction (M) corresponds to 4 μg of chlorophyll and supernatant fractions (S and S3X) correspond to loading of 10 and 30 μg of protein, respectively. Coomassie brilliant blue-stained gel (CBB). The arrow indicates immunoblot signals specific to CPSFL1. (D) Growth phenotypes of C. reinhardtii dark-grown WT, cpsfl1 mutant and cpsfl1 (CPSFL1) complemented line in TAP medium shifted to constant light (65-μmol photons m−2 s−1) for 72 h.