Figure 1.
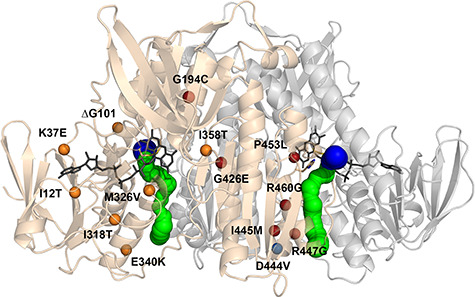
Structure of the hE3 obligate homodimer at 1.75 Å resolution (PDB ID: 6I4Q) with the 14 disease-causing substitution/deletion sites reported to date. The pathogenic substitution/deletion sites are represented in a single hE3 monomer. Spheres designate the Cα atoms of the respective residues and are color coded as follows: variants investigated in the present study—red; variant structure was reported earlier—blue; no variant structure is available—orange. Solvent accessible channels leading to the active sites were computed by Caver applying the dominant Glu332 and Arg460 conformations (see text). The LA-binding and H+/H2O channel segments are blue and green, respectively. Monomers A and B are related by a two-fold symmetry axis and colored beige and grey, respectively. The FAD prosthetic groups, the Cys45-Cys50 disulfide bonds and the His452′ residues are depicted as sticks.
