Figure 9.
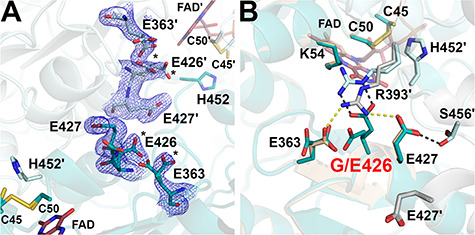
Structure of the G426E-hE3 variant (PDB ID: 6I4U). (A) (2mFo-DFc) composite omit map (1.5σ) confirms the substitution in G426E-hE3. Dominant conformers of Glu426 and Asp363 are labeled with asterisks (*) in both chains. Cys45-Cys50, His452 and FAD are also shown to demonstrate their proximity to the substitution site. (B) Close-up view on the substitution site in G426E-hE3 (dark/light teal; FAD: raspberry) and hE3 (beige/grey; FAD: beige). Disrupted (Glu363-Arg393′, Glu427-Arg393′) and newly-formed (Glu426-Arg393′, Glu427-Ser456′) intermonomeric interactions upon the substitution are represented as yellow and grey dashed lines, respectively.
