Figure 3. Abrogation of NAD+ biosynthesis induces genome-wide disruption of the hepatic circadian transcriptome.
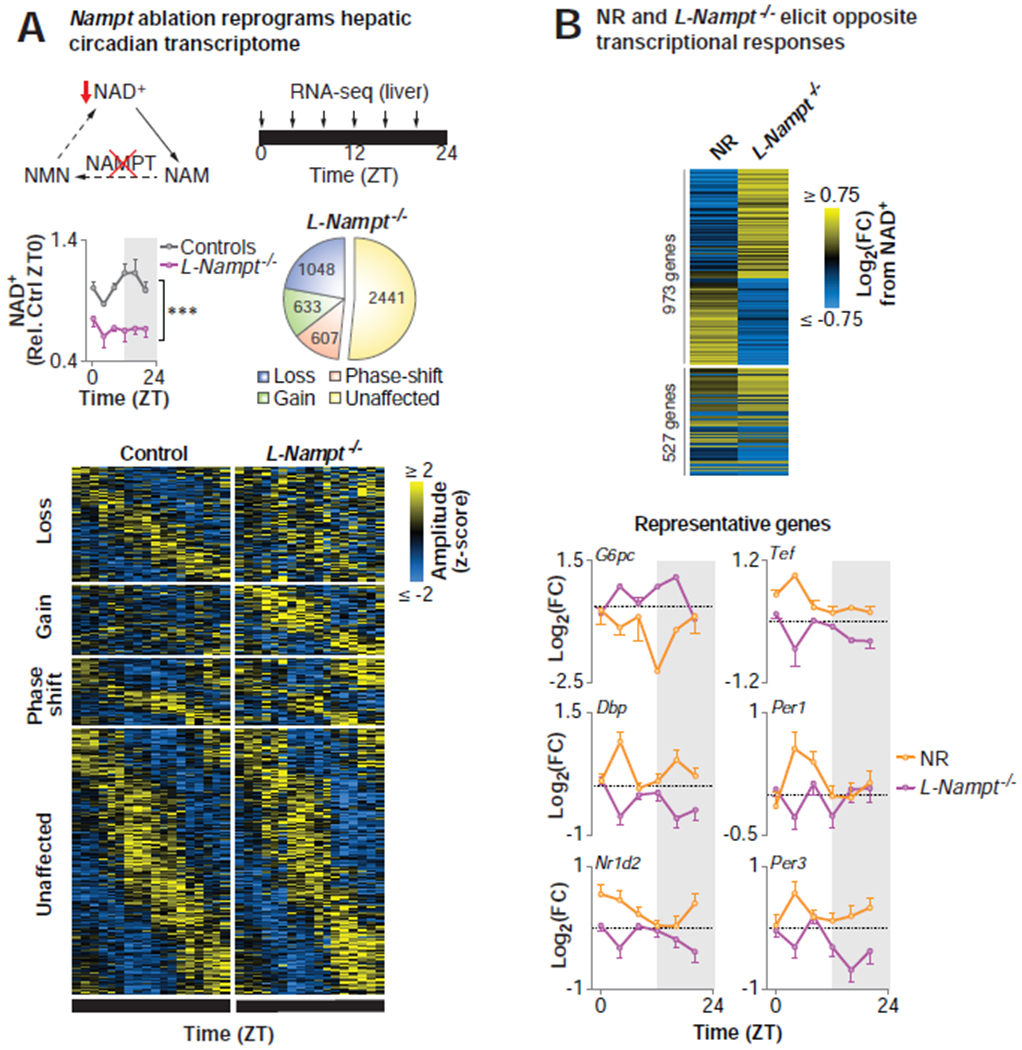
(A) Model depicting the NAD+ salvage pathway, highlighting the contribution of NAMPT to NAD+ biosynthesis. Hepatic NAD+ quantified by HPLC in 4-6 mo old liver-specific Nampt knockout (L-Nampt−/−) and control (Alb-Cre and Namptfx/fx) mice following an 18-hr pre-collection fast (***p<0.001, ANOVA) (n=2-6/timepoint/group). eJTK_Cycle analyses following RNA-seq every 4 hrs across the 24 hr day identified transcripts that lost, gained, or phase-shifted (≥4 hrs) oscillations following Nampt ablation (FDR-adjusted p-value <0.1 and >0.9 for cycling and non-cycling genes, respectively) (n=2-6/timepoint/group). (B) Heat map depicting the maximal diurnal differential transcriptional responses to NR supplementation and NAD+ depletion in L-Nampt−/− mice for the 1500 most up- and down-regulated genes. Log-transformed fold change at each timepoint from NR relative to H2O (orange) and L-Nampt−/− relative to control (purple) for representative genes (n=2-6/timepoint/group). See also Figure S1 and Table S2.
