Figure 3.
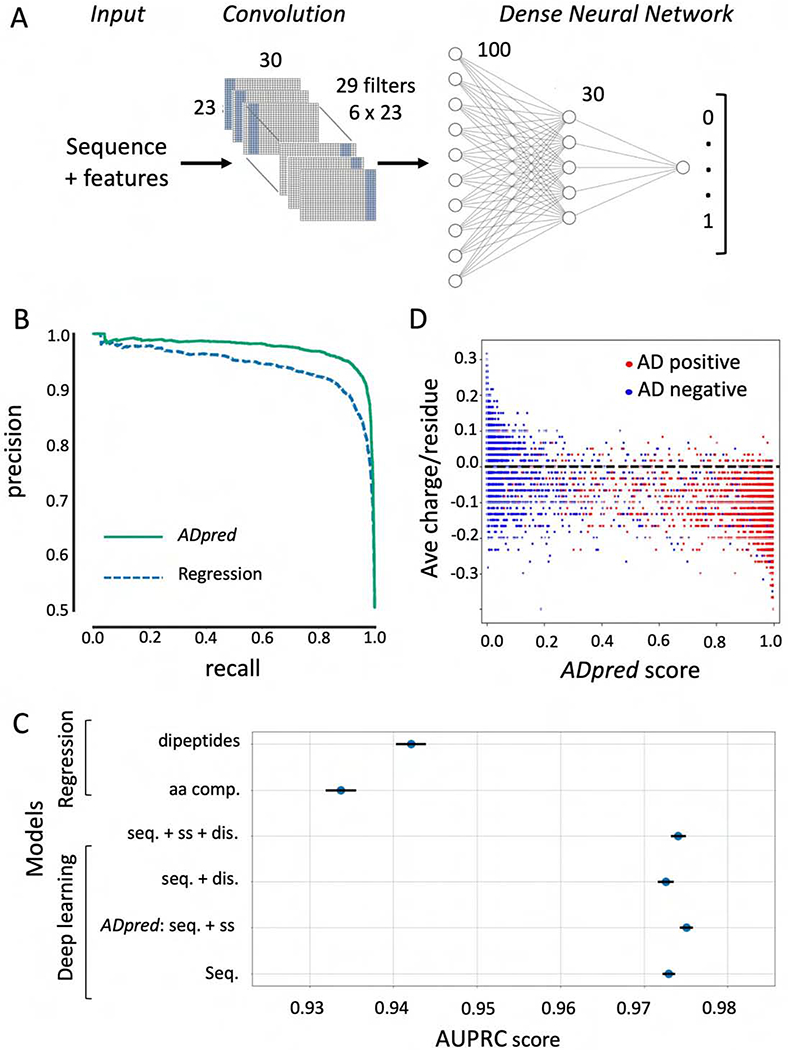
Convolutional deep neural network architecture and performance. A) Input for each sequence consists of the 30 amino acid peptide sequence and its predicted sequence features (secondary structure and/or intrinsic disorder). A convolutional layer learns patterns characteristic of ADs independent of their precise position in the AD sequence. The flattened outcome of the convolution is used as an input for a dense two-layer-network with 100 and 30 neurons respectively. The output layer gives the probability of AD function for the input sequence. B) Analysis of model performance. The precision-recall curve compares the performance of the linear regression model utilizing dipeptide frequencies and the best deep learning model (ADpred) utilizing amino acid sequence and secondary structure predictions. (C) Comparison of several regression and deep learning models evaluated with 10-fold cross validation, with the lines corresponding to standard error of the mean. dis. = disorder predictions; seq. = peptide sequence; ss = secondary structure prediction (statistics from Table S4). D) Correlation between predictions of the deep learning model and the average charge per residue of the 30mers. Dotted line represents peptide with neutral average charge.
