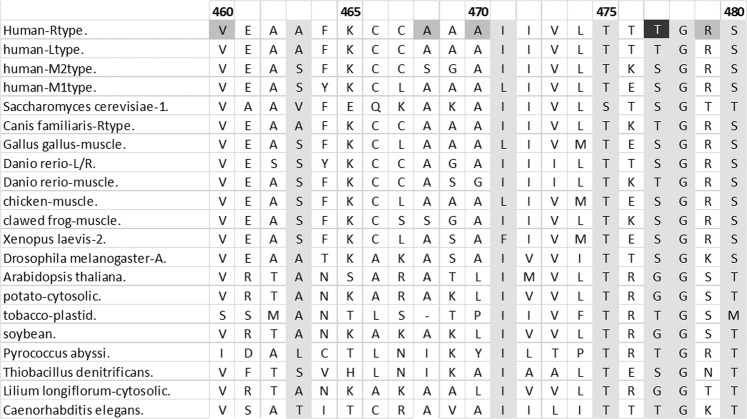
Fig. 1.
An example sequence alignment. Sequence alignments are represented with related protein (homologs) in horizontal rows, aligned so that equivalent positions fall into vertical columns. Amino acids are represented with the one letter code. When the same amino acid is present in many homologs, that position is considered to be conserved. Conservation is interpreted to indicated that other amino acids are not tolerated at that position, which in corollary indicates the importance of that particular side chain. This example shows part of the pyruvate kinase sequence alignment. The four human isozymes are the top rows and position numbering corresponds to RPYK; note that more sequences and more amino acid positions are in the alignment than shown. Light gray columns indicate positions with conserved ConSurf scores in the full alignment (Glaser et al. 2003; Landau et al. 2005; Ashkenazy et al. 2010). Black (conserved) and dark gray (not conserved) cells indicate positions of RPYK disease mutations (Pendergrass et al. 2006)