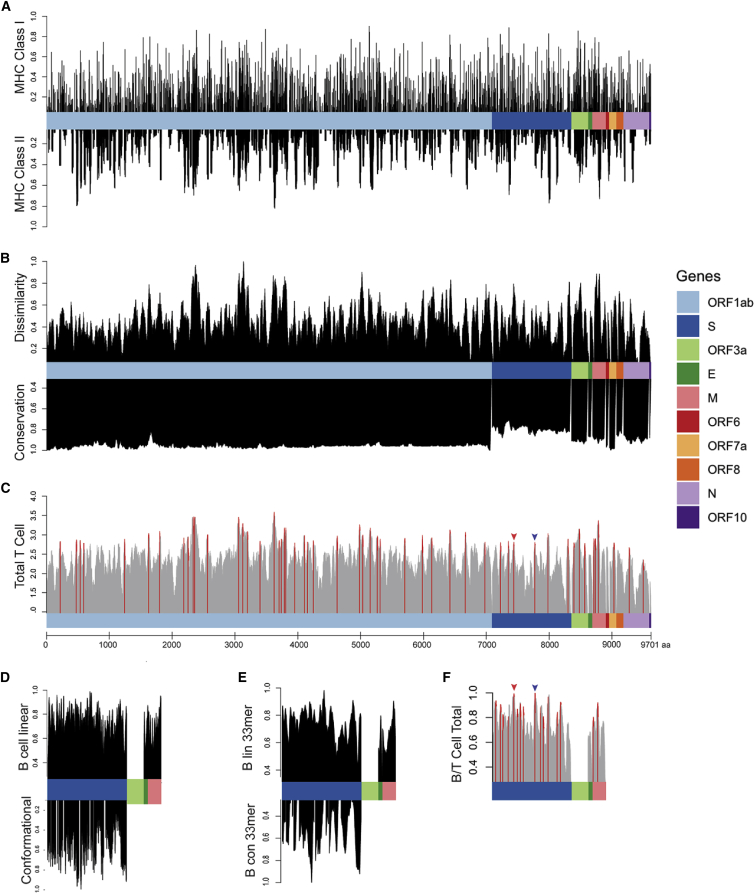
Figure 1.
Epitope Scoring along SARS-CoV-2 Proteome
(A) HLA presentation of 33-mers across viral proteome. Representation of MHC class I presentation (top) and MHC class II presentation (bottom) reported as frequency of the population predicted to present peptides derived from each region of the viral proteome.
(B) Scoring of each epitope derived from the 33-mers along the length of the proteome as compared with the epitopes derived from the normal human proteome presented across 84 HLA alleles, reported as normalized scores in which the highest scoring epitopes are maximally dissimilar to self-peptides derived from normal proteins (top). Scoring for genomic conservation against 15 cross-species coronaviruses and 727 human sequences, with highest scoring regions conserved across human and other mammalian coronaviruses (bottom).
(C) Combined epitope score reported as sum of four above parameters (local maximum for epitopes with 90th percentile total score in red).
(D) Scoring of B cell epitopes for each amino acid for linear epitopes for Spike, Envelope, and Matrix proteins (top) and conformational epitopes in Spike protein (bottom).
(E) Combined scoring of 33-mer epitopes as described in (D).
(F) Combined B and T cell epitope scoring in Spike, Envelope, and Matrix proteins. Receptor binding domain epitope highlighted with red arrow and epitope containing furin cleavage site highlighted with blue arrow (Figure 2).