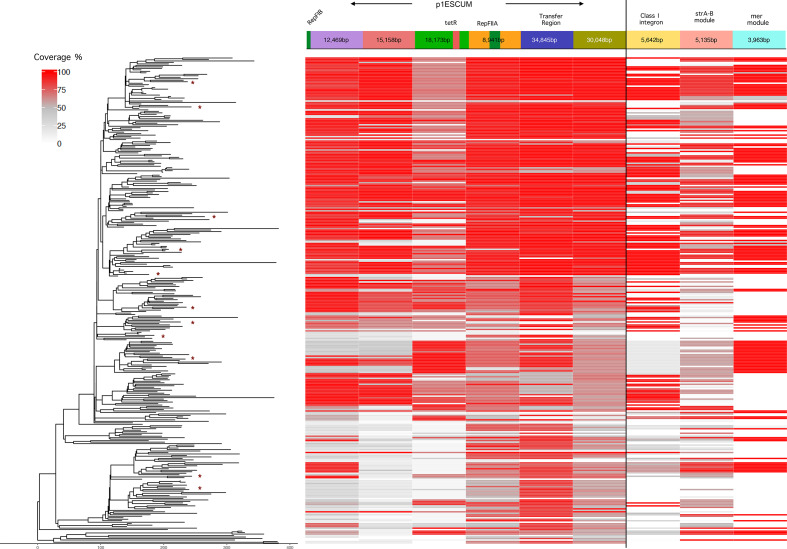
Fig. 4.
Comparison of Global ST69 Isolates. The UMN026 strain was used as reference genome to map the sequencing reads after masking out the mobile genetic regions. The variants were then identified using VarScan and recombinations were filtered by Gubbins. The midpoint rooted phylogenetic tree is built using RAxML. The x-axis of the tree represents the number of base substitutions along the length of the edges of the tree. The * in the tips of the tree indicates 11 of the 24 ST69 Scottish isolates from [31]; the others were much less related to the global isolates and formed an outlier group on the phylogenetic tree and were thus removed for ease of visualisation of the whole ST69 group . The panels to the right show the coverage of reads over p1ESCUM and the various resistance modules indicated to the right.