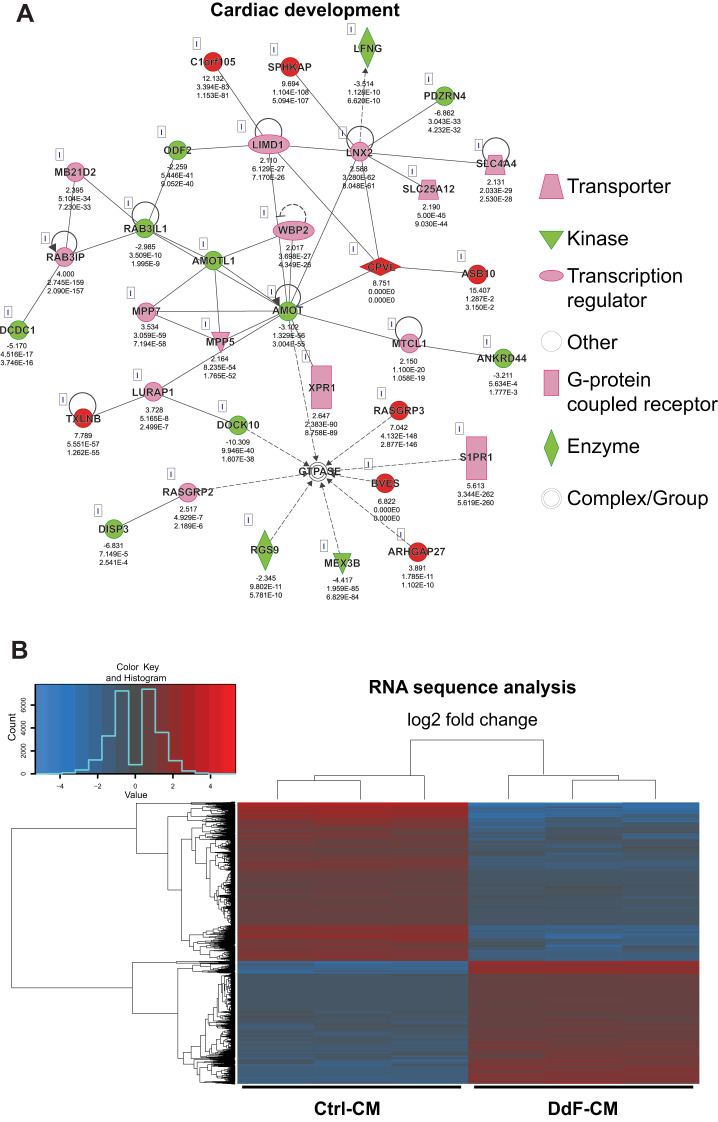
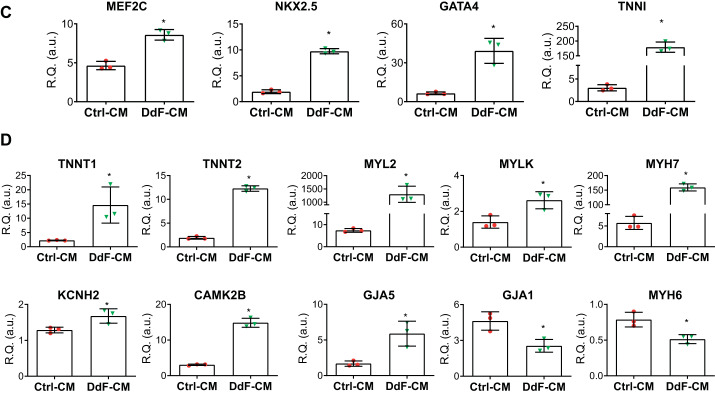
Fig. 5.
Characterization of DNA-damage-free cardiomyocytes (DdF-CMs). RNA sequencing followed by Ingenuity Pathway Analysis was performed on control (Ctrl) CMs and DdF-CMs. A: cardiac development regulatory network in DdF-CMs was upregulated compared with Ctrl-CMs (continuous lines and arrows, direct relationship; broken lines and arrows, indirect relationship). B: heat map of gene expression in Ctrl-CMs and DdF-CMs. The heat map visualizes 2-way hierarchical clustering of 4,245 differentially expressed genes in DdF-CMs compared with Ctrl-CMs (n = 3; P < 0.05, DdF-CM/Ctrl-CM log2 fold change ≥ 2). C and D: transcript expression levels of various differentiation and myocyte maturation markers quantified by RT-PCR in Ctrl-CMs and DdF-CMs. t-test was performed. n = 3. Data are means ± SD. *P < 0.05 vs. Ctrl-CM. R.Q., relative quantity; a.u., arbitrary units.