Abstract
Biosensors-based devices are transforming medical diagnosis of diseases and monitoring of patient signals. The development of smart and automated molecular diagnostic tools equipped with biomedical big data analysis, cloud computing and medical artificial intelligence can be an ideal approach for the detection and monitoring of diseases, precise therapy, and storage of data over the cloud for supportive decisions. This review focused on the use of machine learning approaches for the development of futuristic CRISPR-biosensors based on microchips and the use of Internet of Things for wireless transmission of signals over the cloud for support decision making. The present review also discussed the discovery of CRISPR, its usage as a gene editing tool, and the CRISPR-based biosensors with high sensitivity of Attomolar (10−18M), Femtomolar (10−15M) and Picomolar (10−12M) in comparison to conventional biosensors with sensitivity of nanomolar 10−9M and micromolar 10−3M. Additionally, the review also outlines limitations and open research issues in the current state of CRISPR-based biosensing applications.
Keywords: CRISPR/Cas system, Biosensor, IIoT, Cloud, Big data
Introduction
Currently, the field of smart systems such as smart biosensors, implants and prosthetics are gaining attention due to their function in disease management and control, rehabilitation and other post-surgical operations. Smart biosensors have potential in assisting clinicians while measuring key parameters for monitoring and prediction of diseases for early intervention. Smart sensing and monitoring devices using nano-chips, nano-sensors, nanorobots poses potential in monitoring drug medication, vital signs and detection of pathogens associated with diseases [1, 2]. The artificial intelligence driven biosensor collects physiological and other form of data from patient’s wearable biosensors and applies Artificial Intelligence or Machine learning techniques to detect changes in patients vital signal patterns [3].
The technological advances in the field of communication, data analysis and artificial intelligence have given rise to Internet of things (IoT), Internet of nano Things (IoNT), Industrial Internet of Things (IIoT) and Internet of Medical Things (IoMT). Integration of IIoT in healthcare (IoTH) is transforming the sector in areas such as diagnosis, patient information management, real-time patient monitoring, health management, and medical emergency management. Similarly, the use of Internet of Medical Things (IoMT) as the exchange of medical or healthcare data between people or participant (patients, clinicians, doctors etc.) and medical devices (sensors, monitors, implants etc.) using wireless communication [1].
The use of nanotechnology for design of nanoscale devices such as, nanochips, nanointerface, nanosensor, nano-routers and nano-antennas in different field such as healthcare system, electronics, agriculture etc. stimulates the concept of Internet of nano Things (IoNT). Different studies in biomedicine has shown the promise of nano-biosensor to collect data or information about allergens and pathogens in the environment and the use of body nanobiosensor or nano-implants to collect data based on electrocardiographic signal [50]. Acquiring of data through IoNT will aid in accurate monitoring and diagnosis of patients suffering from different diseases. As the number of IoT connected devices are set to increase rapidly, the use of 5G for data acquired from IoT, IoNT, IoMT devices is highly required to transmit and process the data. This can be achieved as a result of nano-network communication can be in form of nano-molecular, chemical, mechanical and electromagnetic communication [97].
For years scientist have been trying to uncover desired characteristics, systems, mechanism from nature. This led to biomimetics which is a term use for imitations of different models, elements and systems from nature [84]. Since the chain discovery of CRISPR and its components by Ishino, Mojica, Barrangou and Doudna, so many applications of CRISPR/Cas system have been developed for diagnosis and therapy. CRISPR stand for Clustered Regularly Interspaced Short Palindromic Repeat which is a warehouse of arsenics ceased from viruses and stored as a bacterial CRISPR array. Bacteria utilize this array which is made of spacers stored in between sequence of repeats. Each spacer is obtained from different viruses and call to action to fight against matching invading virus. Scientists imitate this mechanism to precisely edit gene of interest that is robust, simple and efficient compare to prior gene editing techniques [7]. CRISPR-based biosensor is a molecular diagnostic tool which utilize a Single Guide RNA (SgRNA) with Cas system such as Cas9, deactivated or mutated Cas9 (dCas9), Cas12a (CPF1) and Cas13a (CPF2) to bind target sequence (dCas9) or cleave target DNA/RNA (Cas9, Cas12a and Cas13a) and generate a signal readout or color change [62].
Among the challenges of application of cloud computing in medical care revolve around data storage, accessibility of data and proper management information [61]. Signals and readout obtained from biosensors are used as data for training using machine learning algorithms. However, with the promise of CRISPR biosensors for point of care application over conventional biosensors, they are still not automated to carry out this task due to limitations regarding fabrication on microchips, signal conversions, wireless transmission and cloud computing. In this review we focused on the potential of artificial intelligence and biomedical big data in developing smart futuristic biosensing paradigms.
Comparison to similar surveys
There have been similar attempts and review in the literature on CRISPR-based biosensors, types, mechanisms, applications, merits and limitations. In this subsection, we overview these surveys and highlights how it contrast with our own review. For example, the most comparable review to our work in terms of CRISPR-based biosensors is given by Li Y et al. [62]. The authors discussed about different classification of CRISPR/Cas biosensing system based on binding and cleavage and Cas effectors. The review also addressed the timeline for development of CRISPR/Cas biosensing technology, example of research articles related to the subject and Pros and Cons of different biosensing approach. Authors have not touched Big Biomedical Data and how data obtain from CRISPR-based biosensors will be stored in Cloud system for analysis, prediction and support decision. Aileni et al. [5] explained the application of Cloud computing for big data acquired from biomedical sensors. Authors concentrated on Cloud Computing architecture, biomedical sensors in general and data analysis but did not pint-point CRISPR-based biosensors and its prospect as the emerging biosensor for diagnosis of diseases.
A related, brief review of CRISPR biosensing as chipping biosensing in diagnostic was addressed by Abudayyeh and Gootenberg [2]. Authors highlighted the application of CRISPR-based biosensors as powerful tools for molecular detection and for point of care application. The review focused on deactivated form of Cas9 (dCas9) paired with gFET technologies for rapid diagnosis of unamplified samples obtained from patient with Duchene Muscular Dystrophy (DMD). In addition, the review discussed on the prospect of CRISPR-based biosensing going digital and how the biosensor can be use in remote areas and in-field tracking of epidemics. Despite this, they did not include other Cas effectors such as Cas12 and Cas13 and the possibility of acquiring data from biosensors for storage in the cloud for big biomedical data and analysis.
Perhaps, the most comparable review to our work in terms of CRISPR were given by Adli [4]. The author discussed about different genome engineering approaches, evolution of CRISPR, the use of CRISPR as a tool for genome engineering and beyond. However, the review did not consider the application of IIoT, cloud computing and big biomedical data for the generation of future biosensors. Kanaparthi et al. 2019 [51] explained the application of different conventional biosensors and nano-based biosensor, the use of artificial intelligence and IoT. However, authors did not explain CRISPR-based biosensor and big biomedical data. Al-Turjman et al. [1] addressed the use of IoT, Biomedical sensors, cloud computing system. Authors did not include CRISPR-based biosensors (Table’s 1, 2, 3 and 4).
Table 1.
Summary of related review
Table 2.
The list of used abbreviations
| Abbreviation | Explanation |
|---|---|
| Am | Attomolar (10−18M) |
| AMIDL | Artificial Muscle Intelligence System with Deep Learning |
| AI | Artificial Intelligence |
| BBD | Big Biomedical Data |
| BP | Base Pair |
| CPF1 & CPF2 | CRISPR from Prevotella and Francisella 1 & 2 |
| CNN | Convolutional neural networks |
| CCS | Cloud Computing System |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeat |
| CRRNA | Clustered Regularly Interspaced Short Palindromic Repeat Ribonucleic Acid |
| CAS-EXARP | CRISPR/Cas9 triggered isothermal exponential amplification reaction |
| dCas9 | Deactivated Cas9 |
| DGMs | Deep Generative Models |
| DMD | Duchene Muscular Dystrophy |
| DNA | Deoxyribonucleic Acid |
| DSDNA | Double Strand Deoxyribonucleic Acid |
| EEG | Electroencephalogram |
| fM | Femtomolar (10−15M) |
| gFET | Graphene-based Feld Effect Transistor |
| GMO | Genetically Modified Organism |
| HEPN | Higher Eukaryotic and Prokaryotic Nucleotide |
| HDR | Homologous Directed Repair |
| HR | Homologous Recombination |
| HSV | Herpes Simplex Virus |
| IIoT | Industrial Internet of Things |
| IoMT | Internet of Medical Things |
| IoNT | Internet of Nano Things |
| IoT | Internet of Things |
| IoTH | Internet of Things in Healthcare |
| ML | Machine Learning |
| NASBA | Nucleic acid sequence-based amplification |
| NASBACC | Nucleic acid sequence-based amplification CRISPR cleavage |
| NGG | Nucleotide Guanine-Guanine |
| NHEJ | Non-Homologous End Joining |
| NPs | Nanoparticles |
| NUC | Nuclease |
| PAM | Protospacer Adjacent Motif |
| PCR | Polymerase Chain Reaction |
| PM | Picomolar (10−12M) |
| PNA | Peptide Nucleic Acid |
| RNA | Ribonucleic Acid |
| RPA | recombinase polymerase amplification |
| RT-RPA | reverse transcript recombinase polymerase amplification |
| SFTS | Severe Fever Thrombocytopenia Syndrome |
| SgRNA | Single Guide RNA |
| SHERLOCK | Specific, high-sensitivity, enzymatic, reporter, unlocking |
| SNP | single nucleotide polymorphism |
| SRSR | Short Regularly Spaced Repeat |
| SSRNA | Single Strand RNA |
| ST | Scrub Typhus |
| SVM | Support Vector Machine |
| TALENS | Transcription Activator Like Effector Nuclease |
| TracrRNA | Transactivating RNA |
| VSV | Vesiculovirus |
| yM | Yoctomolar (10−24M) |
| zM | Zeptomolar (10−21M) |
| ZFN | Zinc Finger Nuclease |
Table 3.
Summary of Conventional Biosensors for detection of pathogens
| Reference | Type/Modification | Biological component | Target/organism |
|---|---|---|---|
| [12] | Electrochemical | RNA | Dengue virus |
| [99] | Optical | Antibody | Herpes simplex virus type (HSV-1) |
| [28] | Electrochemical/miniaturize gold electrode | Antibody | Avian influenza virus |
| [101] | Electrochemical/Silicon nanowire | Peptide nucleic acid (PNA) | Dengue serotype 2 virus |
| [64] | Electrochemical | Antibody | Rotavirus |
| [24] | Optical | Antibody | Vesiculovirus (VSV) |
| [70] | Electrochemical | Bacterial cells | Escheria coli |
| [82] | Microelectromechanical | Immobilized antibodies | Escheria coli |
| [65] | Electrochemical | Bacterial cells | Escheria coli, Paeruginosa, Staphylococcus aureus, Staphylococcus epidermidis |
| [30] | Optical/Quantum dots nanoparticles | Aptamer | Vibrio paragaemolyticus, Salmonella typhimurium |
| [1] | Optical/Graphene magnetic nanosheet | Bacterial cells | Pseudomonas aeruginosa, Staphylococcus aureus |
| [72] | Optical/cationic gold nanoparticle | Enzyme | Escheria coli, streptomyces griseus, Bacillus subtilis |
Table 4.
Summary of CRISPR Based Biosensor
| Reference | Cas Name | Target | Result |
|---|---|---|---|
| [79] | Cas9 | Zika virus | The biosensor was able to discriminate between American- and African-lineages of Zika virus between 2 and 6 h. |
| [44] | Cas9 | DNA methylation and L. monocytogenes | The biosensor detected DNA target within 1 h with sensitivity of 0.82aM |
| [38] | dCas9 | Genes associated with Duchenne muscular dystrophy (DMD) | 1.7fM sensitivity within 15 min |
| [102] | dCas9 | Mycobacterium tuberculosis 165 rRNA | Sensitivity in equimolar |
| [55] | dCas9 | DNA & RNAs of Scrub typus (ST) and severe fever thrombocytopenia syndrome (SFTS) | sensitivity of 0.54aM and 0.63aM in SFTS with less than 20 min for discriminating between ST and SFTS |
| [60] | Cas12a/HOLMES | DNA from cultured human 293 T cells or collected saliva from human individuals | detectable concentration of 0.1 nM without amplification and a 10aM when combine with PCR. HOLMES has shown ability to discriminate single base differences on mutated target DNA |
| [25] | CRISPR/Cas12a | human papillomavirus 16 (HPV-16) and Parvovirus B19 (PB-19) | Sensitivity in Picomolar for detection of HPV-16 and PB-19 |
| [20] | Cas13a | MicroRNAs in blood sample of children with brain cancer | 10pM detection limit with less than 4 h processing time and 9-min readout time |
| [37] | Cas13a (SHERLOCK) | Nucleic acid obtained from Zika virus, dengue virus, bacterial isolates, human DNA genotype etc. | Detecting both RNA and DNA target with single base resolution with attomolar sensitivity. Sensitivity of 2aM in Zika virus |
| [36] | Cas13b (SHERLOCKv2) | RNA | Sensitivity of as low as 2 attomolar |
| [74] | Cas13a (SHERLOCK-HUDSON) | sensitivity of 90aM for detection of Zika virus RNA in serum or whole blood and 20aM in urine with 2 h total turnaround time. Identification of Zika, Dengue, West Nile and Yellow fever viruses |
Other reviews of close comparison are provided in (Batista and Pacheco [15], Eid and Mahfouz [32]). Authors overview Zinc Finger Nuclease (ZFN), Transactivating Activating Like-Effectors Nuclease (TALENS) and CRISPR-based biosensors. However, without the possibility of integrating data obtained from biosensors with IIoT, Big Biomedical Data and Cloud Computing System. Moreover, many review papers on one aspect such as CRISPR in general provided in (Bhaya et al. [17], Doudna and Charpentier [29], Hsu et al. [43], Ishino et al. [45], Makarova et al. [69]) and CRISPR-based biosensors provided in Li and Liu [63].
Scope of this paper
This review paper aims to offer insight into future CRISPR-based biosensor. We overview different aspect of CRISPR-based biosensing. In order to provide the reader with background on CRISPR, first we give insight into history of genome engineering, discovery of CRISPR in nature, the use of CRISPR as a gene editing tool. To relate and compare CRISPR-based biosensor with conventional biosensors, we included some literature on biological component of different biosensor such as enzyme based, antibody-based, nucleic acid-based and cell-based biosensors including modifications with nanoparticles. Moreover, we provide a comprehensive study on CRISPR-based biosensor in general and various classifications such as binding and cleavage biosensors. In each type of biosensor (i.e. conventional and CRISPR-based) we included summary tables to offer an overview on the subject. We delved into IIoT big biomedical data (BBD) and cloud computing system (CCS), Artificial intelligence and their application in healthcare system and the integration of IIoT, BBD and CCS with CRISPR-based biosensor as proposed future biosensor.
In essence, this review aims to integrate the application of IIOT, BBD and CCS with biosensors developed using CRISPR/Cas systems as biological components for generation of future biosensors. It offers detailed description of CRISPR as a gene editing tool and different Cas effectors such as Cas9, Cas12 and Cas13. Also, it opens a window for further research projects for generation of point of care biosensor merged with IIoT, BBD and CCS for diagnostic, analysis, and storage of data.
The remaining parts of this article are organized as follows. Section 2 overviews the application of Internet of Things (IoT), Cloud Computing System, Big Biomedical Data, conversion of biological and biomedical signals to data, DNA-based recording system, Application of Artificial Intelligence in Biosensing Technology and big data from biomedical sensor. In section 3, we discuss about history of genome engineering emphasizing on Recombinant DNA technology, Zinc Finger Nuclease (ZFN), Transcription Activator Like Effector Nuclease (TALENS) and evolution of CRISPR. We explain the concept of CRISPR in nature, application of CRISPR as a gene editing tool and CRISPR database for genome engineering. In section 4, we introduced Conventional biosensors and its classification based on biological recognition elements which include DNA, Antibodies and Enzymes and modifications using nanoparticles. In section 5, we overview CRISPR-based biosensors and classification base on Cas effectors which include Cas 9 (and dCas9), Cas12 and Cas13 while focusing on their mechanism and sensitivity. In section 6, we highlighted few supporting researches and proposed a new biosensing approach that integrates IIoT, Cloud Computing System (CCS) and Big Biomedical Data (BBD) for cloud storage, data analysis, prediction and support decision. In section 7 we outline current open research issues in CRISPR biosensing ranging from low sensitivity, application for point of care diagnosis, in-field tracking, data mining, complexity and privacy of data. Finally, we include Concluding remarks in section 8.
The following table summarizes the used abbreviations all over this document for more readability.
Application of internet of things, big biomedical data and cloud computing system in healthcare systems
Internet of things (IoT)
The emergence of IoT in the last decade has unzip so many applications in different field of science and technology such as environmental sciences, healthcare, engineering and industries. IoT spin around connecting both data or information from sensors embedded within devices such as monitors, mobile phones, sensors to the internet [4]. Industrial Internet of things (IIoT) and Internet of Things (IoT) has now become the ideal and promising approach that can revolutionize and improve conventional healthcare system to e-healthcare systems. In healthcare system, IoT revolve around devices that connect participants such as clinicians, medical doctors, surgeons, nurses and patients and allow exchange of information from one participant to another to achieve efficient and right clinical decisions [50].
Merging IoT with healthcare system will help reduce cost, increase efficiency, enhance sustainability and effectiveness in healthcare such as accuracy and speed [92]. Integration of IoT system with healthcare will link people and devices to a network which is capable of carrying out tasks such as monitoring, diagnosis and remote surgeries over the internet [90]. The concept behind application of IoT in hospitals is to gathered steady streams of data from patient’s implanted sensors, monitoring devices and automatically feed it to electronic medical records for storage and accessibility. Wireless technology in healthcare system will connect all resources which includes clinics, hospitals, rehabilitation centers, pharmacist, medical doctors, nurses, clinicians, nurses, sensors, implanted devices, assistive devices, monitoring devices, ambulances etc. along with patients [33].
Among the first application of IoT in healthcare system is the development of smart rehabilitation to minimize the challenges of scarcity and lack of resources facing the increasing populace of elderly people worldwide [100]. Al-Turjman [11] developed IoT-based detection system for facial disorder using snapped images of infected faces using camera and processing it through the internet. The system combined the mechanism of artificial intelligence that was trained to work as dermatologist that can take facial images as dataset and classified them as infected and non-infected categories.
Application of artificial intelligence and machine learning to healthcare system is on the rise. The use of Deep Generative Models (DGMs) and Convolutional neural networks (CNNs) is utilized to support pathologist in both image and signal analysis and classification for making accurate medical decisions [10]. Jacob et al. [46] adopted artificial intelligence approach to designed Artificial Muscle Intelligence System with Deep Learning (AMIDL) to monitor real time human intentions using Electroencephalogram (EEG) sensors.
Cloud computing
The architecture of cloud computing revolves around collection of services such as infrastructure, platforms, applications and servers that allow data virtualization and storage. Cloud computing can be program to be public, private or hybrid [5]. The trends in IoT which utilize sensors, software, embedded electronics and network connectivity contribute to increase volume of raw data and processing. Storing massive amounts of data requires high parallel power computing and storage [73].
An example on application of cloud computing in healthcare system is Cloud, which is a preventive healthcare approach that analyze physiological signal data and provide early warning on diseases [61]. Collaboration between scientist in the university of Ontario Institute of technology and IBM led to the development of telemedicine based on cloud computing which provide doctors support decision system for remote and rural communities [5].
Protection of data in cloud system is very essential. Thus, computer security as a subbranch of computer science is growing rapidly and it is task on protecting electronic data against attacks (Daniel-of-service), data manipulation, unauthorized access, phishing, hardware theft etc. The main goal of computer security is to protect valuable data, computer hardware, system resources and software. However, data security is focused on protecting data store in cloud systems or transmitted between computer systems. Privacy of data is very critical in cloud computing especially in healthcare sectors. Regulation of access to data and enforcing rules to limit unauthorized individuals from getting access and transmitting acquired data is very important. Integration of data security to cloud system which stored medical data of patients obtained from medical devices, biosensors and implants is highly crucial [18].
Biological/biomedical signals and data
Conversion of biological/biomedical signals to data
Many studies have reported the generation of varying magnitude and order of biotic signals from biological system. These signals can be in form of proteins, nucleic acid, metabolites and ions. Diversity of various signals within a cell contribute to limitation of simultaneously tracking multiple biological reaction over a period of time. Different types of biological data can be obtained from different type of cells in form of spatial, quantity, temporal and identity data. For measurement and analysis of biological activities, RNA, DNA, Protein, inorganic and organic biochemical metabolite forms the molecules of interest [87].
Accurate measurement of biological data from biological process such as biosensing, biomonitoring and biosurveillances has been critical for understanding of complex biological process, interpretation of diagnosis and clinical decision. One of the challenges of measurement of biological and biomedical data include the difficulty of accessing biological environment, organ and tissue (such as brain and gut). Another limitation, is some method lead to cellular or tissue destruction and thus temporal data cannot be accessible. In addition, other method need miniaturization to cellular size to be able to access biological sample [87, 34].
DNA-based recording system
Gene regulation and expression serve as the process that convert biological information encoded in DNA of organisms to actionable cellular response. Apart from DNA that is thought to store information, other biological information storage includes CRISPR-adaptive immunity, mammalian adaptive immunity, phase variation etc. [94]. The high need for robust, less invasive and powerful framework for tracking of both extracellular and intracellular biological or biomedical activities has led to the development of CRISPR nucleases DNA-based cellular recorders. The emerging synthetic biology approach has shown the promise of DNA as a memory device for recording of cellular activities, detection of diverse signals, conversion of signals into DNA alterations, reading and interpretation of signals for analysis and decision. This system can be deployed in live cells for storage of biological or biomedical data into permanent DNA medium for analysis in the future [87].
Big biomedical data
The term big data is a concept used in different field of study such as physical sciences (chemistry, physics, meteorology etc.) finance and accounting (banking and marketing), military and defense sectors and medical sciences (biomedicine and genomics) [73]. Collection of biomedical data is growing exponentially, this multitude data provides significant opportunities to comprehend and also discover the interplay amid various biomedical domains such as clinical data, imaging, biometrics, proteomics, genomics, phenomics and metabolomics. For biomedical sciences, there is a need for developing new technological approaches, software and complex algorithms for collection, management and storage of data [87, 91].
Ma’aSum et al. [68] developed Tele-ECG system for early detection and monitoring of heart disease. The system is designed using 3 different components. The ECG sensors is used to obtained patient heartbeat signal and sent to smartphone via Bluetooth network. Smartphone is employed to both visualize and analyze acquired heartbeat signal and a server which link patient with cardiologist. The Big Data of Discovery Science (BDDS) center is taking vital approach to determine connection between contrasting data sources by mining current database of genomics and proteomics data, clinical assessment and brain images. The integration of these domains will help scientist to uncover new insight and generate new hypothesis [91].
Multimedia application of biomedical sensors and big data analysis
Biomedical sensor is the type of sensor that combine biological and electronic system which collect signals variables from biological component and convert the signals into electrical output. These sensors can be classified into chemical and physical. Chemical biomedical sensors include electrochemical, gas, photometric and bioanalytical. These sensors are utilized to measure concentration of chemical substances or chemical changes. Physical biomedical sensors are utilized to measure physical quantities such as body temperature, blood flow, blood pressure, muscle displacement, skin moisture and bone growth [5].
For big data collection, analysis and storage obtained from biomedical sensors, the data should be subject of decision system [47]. The system architecture comprises of 5 different levels. Level 1 consist of data transmission from biosensors to aggregators. Level 2 is made of big data (data collection, discretization and storage). Level 3 consist of medical information where data are classified and analyze. Level 4 is based on data synthesis (i.e. disease knowledge). Level 5 is all about decision support system based on medical actions. In this context, decision support system is based on the use of algorithms on recursive function which employ stack data structure for data collection. In medical data analysis, decision support system revolves around data mining process which allow data analysis, prediction, classifications, clustering and correlation [5, 16 and 47].
The Multidisciplinary fields consisting of biomedical engineers, clinicians, computer and data scientist have been working together to transform the multimedia application of biosensors in healthcare sectors. Some of the systems include Body Area Network (BAN) which is a network of communicating devices such as actuators and sensors. These devices are either wearable or attached in the body of patients whose signals (i.e. outputs) are processed and transmitted to a server or selected healthcare professionals who can assess the multimedia display of the output in a form of representation signal or graphical output of patients biochemical process (glucose level, pulse rate, heart rate, blood pressure etc.) and environments (temperature, humidity, gases etc.) [50].
Application of artificial intelligence in biosensing technology
The improvement in the field of AI and ML approaches has offer a new dimension in the field of medicine and healthcare. These techniques have been successfully used solving problems in biomedical sciences by using modeling and predictive methods. Clinicians along with computer expert have been working together to developed new method for diagnosis and monitoring of diseases such as cancer, pathogenic diseases, genetic diseases, Cardiovascular diseases etc. [53]. The use of data mining in medicine contribute significantly to understanding of biomedical data, diagnosis and prediction of diseases. The decrease of computational cost and increasing amount of data has allowed different ML algorithms to play vital role in biosensing system for clinical and pathological diagnosis [23].
ML algorithms are classified into supervised, unsupervised and reinforcement. Supervise ML (SML) algorithm is the most common approach utilize in medicine where data are labelled and network or model learn features (for image classification) or identify patterns in data (for prediction) [19]. General techniques used for SML include Support Vector Machine (SVM), Neural Networks (NNs) Random Forest, Decision Tree etc. [78, 97]. Unsupervised ML use unlabeled dataset and model learn to predict outcome based on patterns learned from input data. Basic Unsupervised ML algorithms include clustering and rule mining [27]. The least most utilized ML approach is reinforcement where learning process is based on the use of software to automatically determine the best performance within a specific context [3].
The advances in merging Biosensors with AI has led to development of smart biosensors. Chang et al. [22] developed XPRIZE DeepQ Tricorder biosensor that can diagnose 12 different disorders such as tuberculosis, diabetes, anemia, pneumonia, atrial fibrillation, hepatitis A, urinary tract infection, otitis media, stroke, leukocytosis, chronic obstructive pulmonary disease (COPD) and sleep apnea and capture five real-time vital signs such as respiratory rate, body temperature, oxygen saturation, ECG and blood pressure. Shimzu et al. 2016 applied DL for diagnosis of lung cancer. Nehal et al. [76] applied CNN model for prediction of bacterial contaminations using data obtained from photonic crystal based optical biosensor. Linking of biosensors with Machine learning approaches will enhanced prediction, diagnosis and clinical decision making.
Quan et al. [81] developed artificial intelligence-based system for prediction of hypoglycemia (low blood glucose level). The system is designed using Long-short-term-memory (LSM) and the result has shown high performance of the network in prediction of hypoglycemia after half hour. Marling et al. [71] applied machine learning to improve detection of low blood glucose level from data acquired from noninvasive sensors. Dataset include heart rate, skin air temperature and galvanic skin response. The system utilized SVM to train and classify data into hypoglycemic and hyperglycemic.
History of genome engineering
Eukaryotic organisms are composed if billions of DNA bases (Adenine, Guanine, Cytosine and Thymine) that made up their genomes. In molecular biology and medicine, one of the most sought-out goal is for scientist to be able to edit these DNA bases precisely and accurately at a target sites without destruction that can lead to mutations. Physicians have been trying to cure genetic diseases such as sickle cell anemia, cystic fibrosis, Huntington’s disease, αand β thalassemia etc. and to developed a sensitive molecular diagnostic tool that can detect pathogens at a very low quantity. The glimpse of this hope suffices to light in the 1970s as a result of discovery of restriction enzyme and recombinant DNA technology [39]. This mechanism was utilized by scientist to edit and manipulate genome of organisms in a test tube but encountered so many challenges such as resistance in expressing new genes, cell stress responses activation, posttranslational modifications, low solubility etc. [4]. The discovery of homologous recombination (HR) by Capecchi and Smithies has shown the incorporation of exogenous copy of DNA into mammalian cells. Like Recombinant DNA technology, it has its own limitations such as high off-target rates [4].
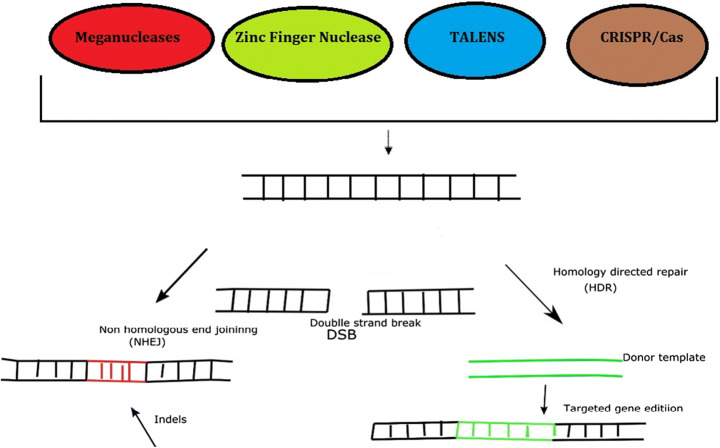
The discovery of Zinc Finger Nuclease (ZFN) opens the window to a new gene editing technology. ZFN utilize small protein motifs that are regulated by zinc-ion which bind to target DNA in a sequence specific manner as each module is able to recognize a 3 base pair DNA sequence [29, 67]. The integration of Zinc Finger proteins with FOK I endonuclease increase efficiency. To this achievement scientist began to engineer genomes of living organisms (both prokaryotes and eukaryotes) and employ this approach for therapy [21, 77]. Instead of recognizing 3-bp DNA sequence, a new discovered gene editing tool known as TALENS (Transcription Activator Like Effector Nuclease) recognize 1-bp which offer better efficiency than ZFN but still has its own limitations [88]. The main principle of ZFN, TALENS is shown in Fig. 1.
Fig. 1.
The main principle of common Genome editing tools
Evolution of CRISPR
The discovery of CRISPR transformed genetic engineering as it offers wide application in medicine and biotechnology. Unlike ZFN and TALENS, CRISPR is very simple to use, cheap and offer better efficiency than prior gene editing tools. It all started in 1987 during and experiment of gene responsible for isozyme conversion to alkaline in Escheria coli by Japanese scientist who discovered a unique and palindromic sequence which read identically both forward and backward direction but the function of the sequence was not known to them. In 2002 the sequence was name Short Regularly Spaced Repeat (SRSR) and rename CRISPR in 2002 by Jansen. Another recipe known as Cas genes was discovered by a group of scientists, these genes are translated to proteins known as CRISPR associated system (Cas) which has nuclease domains and form complex with guide RNA to locate and cleaved target DNA sequence [13]. It was not clear until between 2005 to 2007 that Mojica discovered same sequence in archaea.
The function of CRISPR remained unknown until a research by Barrangou and Horvath who discovered that CRISPR is an immune mechanism employ by bacteria and archaea against invading viruses [14, 66]. The most important component that will pave a way for CRISPR as a gene editing tool was discovered by Charpentier and Doudna who create artificial single guide RNA (SgRNA) which is a chimeric RNA that is made of CRISPR RNA (CrRNA) and Transactivating RNA (TracrRNA). Different studies have shown the efficiency of CRISPR/Cas system to edit microorganisms. The first application of CRISPR/Cas system to edit mammalian cells is carried out by Feng Zeng [14, 45].
CRISPR in nature
CRISPR is an adaptive immune mechanism adopted by bacteria and archaea to fight viruses [42, 66]. Viral DNA is detrimental to bacteria, to prevent viruses from replicating inside bacterial archaeal cells, these microbes utilizes three steps process namely adaptation, recognition (expression or biogenesis) and interference as shown [17, 35, and 49].
Adaptation occurs when bacterial or archaeal cell first came in contact with viral DNA, the CRISPR array translated Cas genes to Cas proteins (Cas1,2 and 9) which locate the PAM sequence in the viral DNA and cleave part of it and store it at the leader strand of the CRISPR array. Recognition stage occurs when the viral DNA attack for the second time, the host’s CRISPR array transcribed it stored spacers into Pre-CRISPR RNA (which are small non-coding RNA) and base pair (link up) with TracrRNA to form a matured CRISPR RNA (CrRNA). In interference step, Cas 9 protein which are translated from Cas genes adjacent to loci form complex with CrRNA and locate the viral DNA through PAM (i.e. NGG) and cleaved matching viral DNA leading to immunity) [14, 17, 35, 42].
Crispr as a gene editing tool
Biomimetics or biomimicry has been utilized by scientist for approximately 3.8 billion years as a principle or approach of emulating nature’s systems, models, strategies and materials to solve human problems [84]. The discovery of CRISPR/Cas system in different species of bacteria and archaea has led researchers to mimic the same mechanism and principle to deliberately edit (insert or delete) genes in both prokaryotic and eukaryotic organisms and is term as “Genetic engineering” [29, 67]. Genetic engineering has witnessed transformation from the discovery of restriction enzymes, recombinant DNA technology, ZFN and TALENS which are very complex, expensive and adversity for multiple edition to the current CRISPR Cas system which is robust, simple, cheap and can target multiple site or target with high efficiency [66, 88]. CRISPR/Cas system approach has taken over as the most reliable and utilized gene editing tool due to its simplicity, accuracy, precision and its ability to target genes at multiple sites over prior tools. There are different class of CRISPR Cas systems such as Cas9, Cas12a, Cas13a and Cas 14 but Cas9 is the most utilized class use for gene editing.
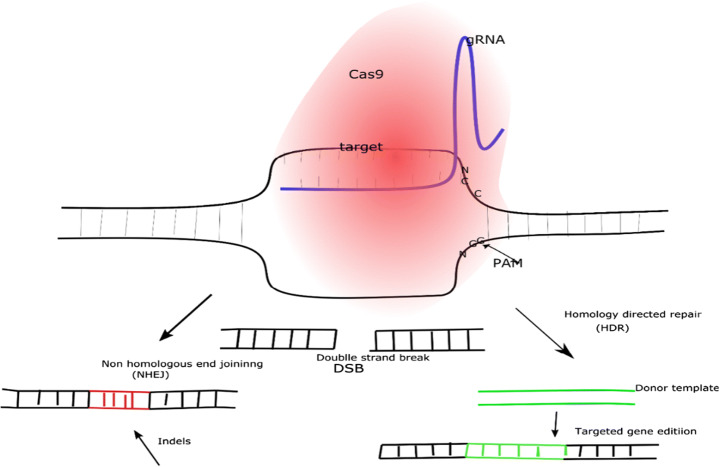
CRISPR/Cas9 is a complex structure that is made of Cas9 enzyme which poses endonuclease domain and single guide RNA (SgRNA) which compose of fusion of CrRNA and TracrRNA. SgRNA is a synthetic form of RNA designed by scientist which contain approximately 100 nucleotides, to navigate the target sequence, the first 20 nucleotides guide the SgRNA matching target sequence. Different bacterial species are used by scientist to isolate Cas9, some of these bacteria are Staphylococcus aureus, Streptococcus pyogenes, Streptococcus thermophilus and Brevibacillus laterosporus [29, 67]. Storage of CRISPR sequence in the form of data storage for DNA with multiple pairs as unit of memory bit (i.e. 100 bp, 20 bp, 500 bp etc.) is now possible [87]. Currently, there are various database and website that provide users with target sequence, gRNA design with optimal off-target.
Scientist took advantage of the two domains present in Cas9 which include RubC and HNH domains; RubC domain cleaved the opposite strand of target DNA while HNH domain cleaved complementary strand [67]. After cleavage, cells utilize Non-Homologous enjoining (NHEJ) and Homology Directed Repair (HDR) as shown in Fig. 2. NHEJ is the most utilized repair mechanisms employed by cells which include addition of deletion of nucleotides known as indels, doubles strand break is repair by either sticking the damage strand together or synthesizing new nucleotide to repair damage (as shown in Fig. 2) which make this mechanism prone to mutations. To study functions of genes, scientist employ NHEJ where gene is disrupted to deliberately stop protein formation [32, 67]. HDR is the most efficient repair mechanism which involve replacing a gene (gene insertion) or DNA sequence with newly synthesize strand. Scientist utilize the newly strand known as homologous donor which undergoes modification (insertion or deletion of nucleotide) and is used by the cells to repair the damage [29, 48, and 98].
Fig. 2.
Genetic Engineering using CRISPR
CRISPR-database for genome engineering
The mechanism behind the use of CRISPR as a tool for genome engineering rely upon the use of guide RNA which contain specific 20 base pair (bp) target sequence adjacent to Protospacer adjacent motif (PAM). Cas system can be re-programed by replacing the first 20 bp with another desired guide and function the same way by locating target and inducing double or single strand break at PAM site of the target genome [43]. Currently, there are many databases for selection of target and designing of guide RNA. Some of the database require input of gRNA sequence while others provide basic genome browser and scores for off-target.
Hodgkins et al. [41] developed Wellocome Trust Sanger Institute Genome Editing Database (WGE) which compute, visualize and select optimal CRISPR sites in a genome browser website. The database not only stored single and paired CRISPR sites but also pre-calculation of CRISPR-off target in both human and mouse exomes. Rauscher et al. [83] developed a database called GenomeCRISPR which contain over 500 thousand Single guide RNAs (SgRNAs) derived from 48 human cell lines in 84 different experiments. The database provides options for data mining and tools that include search of gene or region in a particular genome, track view of Phenotypic and genome of different screens and the impact of various SgRNAs on target gene.
Park and Bae [80] developed an online tool, Cas database and gRNA library of Cas9 nuclease derived from Streptococcus pyogenes (SpCas9). The database provides target sequence with optimal off-target and programming interface (Web Api) for users interested in advanced bioinformatics. Lenoir et al. [59] developed Pooled In vitro CRISPR knockout library Essentiality Screens (PICKLES) which allow users to visualize and download raw or normalized essentiality profiles from 18 thousand plus protein-coding genes across over 50 cell lines. Presently, there are multiple database available online which provide researchers with options of target sequence selection, gRNA design and off target score.
Conventional biosensors
Biosensors are defined as analytical tools which incorporate biological component such as cells, tissues, organelles, microbes, antibodies, cell receptors, Nucleic acid, enzymes or biologically derived and modified materials such as engineered aptamers and proteins, recombinant antibodies and biomimetic (such as imprinted polymers, synthetic catalyst, combinatorial ligands etc.) integrated in physiochemical transducers which may be magnetic, electrochemical, piezoelectric, optical, micromechanical and thermometric. Biosensors employed 3 major biological recognition element which includes Nucleic Acid (DNA and RNA), Antibodies (recombinant, monoclonal and polyclonal) and enzymes [58, 75]. Some of the characteristics of ideal biosensor include versatility, high specificity (i.e. response towards analyte), shorter recuperation time, delicacy, ability to catch and identify low levels of the analyte and high recurrence [89].
Prior to modification of CRISPR for use as biosensors, scientist utilize benchwork diagnosis assays which require bulk procedures accompanied by reagent treatment, tedious sample, sophisticated tools and well-trained laboratory technologies or clinicians. Wide range of biosensors were developed for detection of pathogens such as bacteria, fungi, viruses etc. PCR was developed in 1980s which is primarily use for amplification of Nucleic acid. This technique is utilized to detect target bacterial DNA through amplification and quantification. Different PCR method such as real-time PCR, multiplex PCR and reverse transcript PCR were developed for bacterial detection [58]. Clinicians rely mostly on Immunology based method such as antigen-based approaches which has limitations in terms of results such as low sensitivity, specificity and time consuming.
To enhanced and modified conventional method of diagnosis, scientist developed nanoparticle-based biosensors. These biosensors are incorporated with nanoparticles (NPs) for more reliable, flexible and sensitive diagnosis. Different types of nanoparticles with desired characteristics are employed such as optical (Gold NPs, Cdse/ZnS core/shell dendron, dendrimers, dye-doped silica NP, liposome etc.), magnetic (paramagnetic, supermagnetic), electrochemical (single walled Carbon nanotubes, Gold NPs modified on screen printed carbon electrodes, Au NP based graphite epoxy composite, Platinum NP coated nanoporous film et.) for detection of wide range of pathogenic bacteria such as Salmonella, S. aureus, B. anthracis [95].
CRISPR-based biosensors
The discovery of CRISPR as an adaptive immune system in bacteria and archaea has led to revolution for its application as a programmable gene editing tool for manipulation of both RNA and DNA for treatment of diseases and for studying gene functions and proteins. CRISPR/Cas system has emerged as sensitive and specific molecular diagnostic tool for detection of diseases and for use as a point care diagnosis which include genotyping and clinical diagnostic. This system is highly specific due to hybridization of matching sequence, sensitive, rapid, cheap and very simple to use [2].
Development of reliable techniques for selective and rapid detection of pathogens are critical for diagnosis of infectious diseases. The discovery of CRISPR/Cas system has open ways to many applications. This approach is not solely use as genetic tool for editing prokaryotic and eukaryotic genes to create genetically modified organisms (GMO) and therapies against genetic diseases, recently, scientist harnessed the cleavage and binding activities of this system to developed a novel biosensor with high accuracy and specificity for identification of pathogens compared with conventional biosensors. Some of these tools can only identify target sequence while others can identify and cleave target through navigation of designed guide RNA and Cas system [93]. To better this technique, scientist have integrated CRISPR/Cas system with machine learning to minimize off-targets effect. CRISPR-based biosensors can be divided based on binding or cleavage and based on Cas effectors (Cas9/dCas9, Cas12 and Cas13) [62]. In this article, we concentrated on classification based on Cas effectors.
CRISPR/Cas9 and dCas9-based biosensors
CRISPR/Cas system derived from S. pyogenes is the most utilize type II Cas9 system employed by scientist due to its simplicity and precision. The system utilizes a single guide RNA (SgRNA) to recognize and cleave target DNA. SgRNA is designed with 100 nucleotides but the specificity lies within the first 20 nucleotide which can detect the target through PAM sequence (i.e. NGG). The specificity of these system allow scientist to developed diagnostic tool for virtually any target gene of interest in a precise and quick manner. The system can be programmed to target any gene by simply changing the guide RNA which are now available at various CRISPR database. This property makes it easy to distinguish between strains of organisms which are genetically and morphologically related to each other (i.e. single nucleotide polymorphism) [93].
Amplification of Nucleic acid increase the copies of target for efficient hybridization. Collins and Co-workers developed a sensor that utilize CRISPR/Cas 9 with amplified DNA target using Nucleic acid-sequence based amplification to discriminate viral strains with a single base difference but the research was limited due to requirement of PAM to cleave DNA sequence which limit it signal amplification and for use as a diagnostic tool [103]. Recently Cas9 has shown its “Magic power” for use in a biosensor. This technique employed the cleavage activity of Cas9 and amplification of Nucleic acid as shown in Nucleic acid sequence-based amplification (NASBA), Nucleic acid sequence-based amplification CRISPR cleavage (NASBACC), CRISPR/Cas9 triggered isothermal exponential amplification reaction (CAS-EXARP) which are able to genotype pathogens to an extend of differentiating strains based on single nucleotide polymorphism (SNP) [58].
Sensitive and specific detection of biomolecules at a lower concentration without undergoing amplification is highly required for diagnosis of pathogens associated with diseases. Koo et al. [55] developed a biosensor made up of CRISPR deactivated Cas9 (dCas9) integrated with single microring resonator which utilize DNA and RNA for detection of tick-borne illness which include scrub typhus (ST) and severe fever thrombocytopenia syndrome (SFTS). The biosensor was able to detect ST with a sensitivity of 0.54aM and 0.63aM in SFTS and was able to distinguish between SFTS and ST in blood serum sample in less than 20 min.
Graphene as a nanomaterial poses unique and required properties such as chemical stability in salty water, high electron mobility, these properties makes it the best and noble nanomaterial with an electrical conduit for reporting the activity of the employed biological machineries. Hajian et al. [38] developed a CRISPR-chip based on deactivated or mutated CRISPR/cas9 (dCas9) integrated with graphene-based field-effect transistor (gFET) for detection of genes associated with Duchenne Muscular Dystrophy (DMD) without amplification. A chemical linker is use to link neutered Cas9 enzyme on graphene and the binding of this bio-robot (CRISPR-chip) with target will result in change of energy which is sense on the surface of the graphene. The result is obtained due to changes in the sensing current between source and drain electrode as a result of hybridization of matching sequence (binding) using a commercial reader leading to a sensitivity of 1.7fM.
Pardee et al. [79] developed a paper-based optical CRISPR/Cas9 biosensor for the detection of Zika virus at femtomolar (fM) concentration. The sensor was developed as a result of integration of CRISPR/Cas9 with optical Geno-biosensor which make it highly sensitive and specific due to hybridization of gRNA and Zika virus RNA. Dengue virus was used as a negative control and the biosensor was able to discriminate between different viral strains with single base polymorphisms.
Haung et al. [44] developed a biosensor which combine the binding and cleavage of CRISPR/Cas9 with target isothermal DNA amplification using exponential amplification (CAS-EXPAR). The sensor utilized fluorescence monitoring method to target amplified DNA fragment as a result of cleavage by Cas9. The system was able to detect DNA target within 1 h with sensitivity of 0.82aM and displayed high specificity in discriminating between single base mismatches. Zhang et al. [102] developed an in vitro detection of nucleic acid sequences in Mycobacterium tuberculosis. The system is designed using pair of dCas9 linked to luciferase which is an oxidative enzyme that produced bioluminescence. 2 dCas9 were employed to bind PCR-amplified DNA target to induced spatial overlap of different fluorescence labels (colocalization). The system was able to detect target gene (Mtb 165 rRNA) with a sensitivity of equimolar.
CRISPR/Cas12a-based biosensors
Cas 12 belong to class 2 type V-A, also known Cpf1 is an acronym for CRISPR obtained from Prevotella and Francisella 1. Cas 12 is commonly known for its single break on target sequence due to presence of only RuvC domain (i.e. without HNH domain like Cas9) which induced staggered cut on target DNA. Unlike other Cas system, Cas12 does not require transactivating RNA (TracrRNA) and recognize T-rich PAM sequence [56, 69].
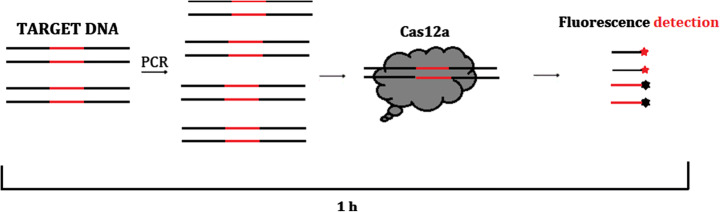
The field of CRISPR-based biosensing is growing but the current biosensors are still not able to match the require simplicity, sensitivity, specificity, cost and speed. Li et al. [60] designed a CRISPR-based detection method which employ Cas12a and a probe (HEX-N12-BHQ1) a quenched fluorescent SSDNA reporter to developed Nucleic acid detection method known as HOLMES (one hour low-cost multipurpose highly efficient System). This system is multipurpose due to its ability to detect target DNA. The mechanism of this approach relies on the present of target DNA/RNA in the reaction system which forms a ternary complex with Cas12a CrRNA binary complex and illuminating of the HEX-fluorescence as a result of trans-cleavage of non-targeted SSDNA reporter as shown in Fig. 3.
Fig. 3.
Detection of target and non-target DNA using HOLMES
Cas12a derived from Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a), was employed as Cas system for HOLMES due to it suitability (compare to other 11 derived Cas12a). the result has shown a detectable concentration of 0.1 nM without amplification and a lower concentration of less than 10aM is achieved when combine with PCR. HOLMES has also shown ability to discriminate single base differences on mutated target DNA (after inducing mutation at different position on target DNA sequence). In order to achieve portable and cost-effective biosensor, Dai et al. [25] developed a universal electrochemical biosensor that utilize Trans-cleavage activity of CRISPR/Cas12a derived from Acidaminococcus sp. (AsCas12a) and Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a), [15]. The biosensor was employed to detect parvovirus B19 (PB-19) and human papillomavirus 16 (HPV-16) at limit of detection (LOD) of 50pM sensitivity. For detection of TGF-β1 (Transforming growth factor) an aptamer-based E-CRISPR cascade was designed.
CRISPR/Cas13a-based biosensors
A comprehensive study of type VI CRISPR/Cas system in 2016 has led to discovery of C2C2 (Cas 13). Unlike Cas9, Cas13 poses no detectable RubC domain, instead it poses 2 higher Eukaryotic and Prokaryotic Nucleotide (HEPN) binding domain. These domain poses RNA activity. Cas 13 hunt for target RNA using RNA guide and when it locates matching target become activated and cleave target and, in some circumstances, it cuts any RNA it encounters a process termed as Collateral cleavage. Collateral activity was harnessed to cut labelled RNA reporters for detection of target Nucleic acid which may come from Bacteria, viruses or eukaryotic cells. It cleaves not only DSDNA but SSRNA and uses single CRISPR RNA as it guides. Cas13a from Leptotrichia wadei (LwaCas13a) where the crRNA–Cas13a complex has a bi-lobed architecture, consisting of a nuclease (NUC) lobe and a crRNA recognition (REC) lobe [3, 62].
Scientist harnessed this unique Cas13 cleavage collateral cleavage activity to cleaved labelled RNA-reporters in order to detect target nucleic acid from both prokaryotic and eukaryotic cells [31]. Nowadays, clinicians employed MicroRNAs (which are non-coding RNAs) as biomarkers for detection of diseases. In order to detect disease at early stage and to avoid progression. Bruch et al. [20] developed electrochemical microfluid biosensor with CRISPR/Cas13a without amplification for onsite detection of microRNAs (miR-lab) in children’ s blood serum with brain cancer. The biosensor was able to detect tumor markers microRNA with a detection limit of 10 pm with overall process time with less than 4 h and readout of 9 min.
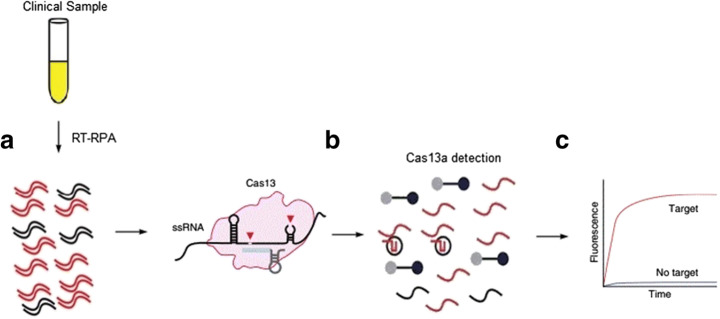
Gootenberg et al. [37] developed SHERLOCK (Specific, high-sensitivity, enzymatic, reporter, unlocking) coupled with recombinase polymerase amplification (RPA) or reverse transcript recombinase polymerase amplification (RT-RPA) and T7 transcription. The biosensing system harnessed the collateral cleavage activity of Cas13a derived from Leptotrichia wadei (LwCas13a) for detection of nucleic acid. SHERLOCK has shown efficiency in detecting both RNA and DNA target with single base resolution with attomolar sensitivity. The system has been utilized to detect bacterial isolates, dengue virus, Zika virus, cancer mutations, human DNA genotype, and antibiotic resistant gene. A new version SHERLOCK is also developed by Gootenberg et al. [36] which is known as SHERLOCKv2 which utilized Cas13b instead of Cas13a coupled with Csm 6 to increase sensitivity by 3.5-fold. Cas13b is RNA-guided RNase CRISPR from type VI-B. like Cas13a, Cas13b derived from Prevotella sp. (PsmCas13b) also possess collateral cleavage activity against target RNA with a sensitivity of as low as 2 attomolar. The system has been utilized to detect dengue virus, Zika virus and mutations in patient liquid biopsy samples.
Myhrvold et al. [74] developed a field deployable viral diagnostic system which combine SHERLOCK (Specific High-sensitive Enzymatic Reporter Unlocking) with HUDSON (Heating Unextracted Diagnostics Samples to Obliterate Nuclease) method which used chemical reaction and heat in bodily fluid to lyse viral particles and inactivate high level of ribonuclease for detection of viruses from bodily fluids. The research utilized samples obtained from 2015 to 2016 pandemic. The mechanism of Hudson and SHERLOCK are shown in Fig. 4.
Fig. 4.
SHERLOCK-HUDSON detection of Viral DNA
SHERLOCK-HUDSON achieved detection in patient sample within 2 h and SHERLOCK detect both dengue and Zika virus as low as 1 copy per microliter concentration. SHERLOCK-HUDSON achieved sensitivity of 90aM for detection of Zika virus RNA in serum or whole blood and 20aM in urine with 2 h total turnaround time.
Validation
Validation of result is very critical for the use of CRISPR-based biosensors for clinical detection of samples and gene editing techniques. Mostly, studies have reported off-target as one of the major limitations of CRISPR-gene editing approach. However, studies have shown the possibility of on-target genomic damage. These damages occur or near the point or at the point where CRISPR is employed for gene editing. To fully utilized the application of CRISPR-toolbox, it is critical to address the stated limitations in order to avoid deleterious effects and to design robust approaches for evaluation of results. Scientist employed different validation process. One of the approached is the used of Single-Molecule Real-Time (SMRT) sequencing technology. This technology has shown efficiency in characterizing and detecting the effect of CRISPR gene engineering experiments. SMRT can now be employed by scientist to verify results of CRISPR experiment before moving to the net step [40, 57]. The use of control samples is another strategy for validation of results where the use of CRISPR/Cas system is tasted on control specimen to determine the biosensing, sensitivity and specificity of the approach. Dai et al. 2019 evaluated the generality of Cas12a designed to detect HPV by detecting ssDNA of erthrovirus and Parvovirus B19 (PB-19).
The sensor developed by Bruch et al. [20] was validated by detecting target using quantitative real-time polymerase chain reaction method in relevant clinical specimens (such as serum) at low concentrations. The validation process was tested on specimens obtained from patient suffering from medulloblastoma. Hajian et al. 2019 evaluated the binding specificity of CRISPR Chip to DSDNA by using PCR product of blue fluorescent gene to validated the gene editing technique. The Cas12a CRISPR employed by Li et al. [60] was validated by using primers for PCR amplification designed with PAM sequence (due to the fact that there may be absence of suitable PAM sequence near the SNP site). However, this method allowed detection of SNP that exist among target genes.
Proposed biosensor
Supporting research and articles
Big data information collected from monitoring of patient’s health using biomedical sensors require algorithm optimization and high computing power to reduce energy consumption that could be generated by transmission of big data in future [5]. One of the challenges of cloud computing application in healthcare system is tune around data storage, accessibility and management of health information [61]. The entire power platform of cloud computing is underutilized and thus the use of biomedical data, IoT and cloud computing will open a gateway and form a basis on our proposed CRISPR-based biosensor. CRISPR-based biosensors have shown promise for use as a biosensor for point of care application with equal or superior performance compare to conventional biosensors. These biosensors have displayed a great potential for automated diagnostic approach, precise therapy, biomedical big data analysis and medical artificial intelligence [62].
Hajian et al. [38] and Koo et al. [55] has shown the potential of CRISPR-based chip going digital. These studies have demonstrated the feasibility of utilizing CRISPR-based biosensors for detection of DNA and RNA associated with disease in healthcare system. Even though there are still some challenges that must be overcome such as purification of DNA steps which is required and critical for compatibility with unpurified DNA samples which hinders its application as a point of care biosensor. Other challenges are the utilization of these sensors in other sample matrix such as saliva and serum and fabrication of portable system for field ready diagnosis [2].
In addition to the characteristics of ideal biosensor listed in section 4, biosensors must be developed to achieved desired aim for detection of diseases and most be able to wirelessly transmit biomedical data to a designated device or database. Moreover, the ability of biosensors to generate vast amount of data for therapeutics contribute to their capabilities for real-time decision making [89]. The use of biosensor for storing of data in cloud system which can be freely accessible by scientist for data analysis and prediction using AI will make it ideal for futuristic biosensor. Merging AI and ML technologies with biosensor can be used to develop POC diagnosis kit that can be deploy for used in remote communities that lack basic facilities [95]. However, development of biosensor enabled with AI will serve as the backbone of next generation biosensors that can be used for real-time, POC and monitoring of diseases.
Solving above challenges will relinquish CRISPR-based biosensor as the most accurate, sensitive, specific and robust point of care diagnostic tool for detection of diseases [2]. A noble CRISPR based biosensor is one that does not require amplification (i.e. using PCR, isothermal Nucleic acid amplification etc.), inexpensive, single molecule sensitivity, single molecule specificity, high speed, portable and can be used as point care diagnosis [103]. Integrating CRISPR/Cas system with nanomaterial that has required properties such as chemical stability in salty water, high electron mobility, biocompatibility and electrical conduit for reporting the activity of the employed biological machineries. For designing of advanced biosensors, the use of nanotechnology is highly recommended due to its ability to enhance selectivity, sensitivity and lower cost of diagnosis [86].
Architecture and framework of proposed biosensor
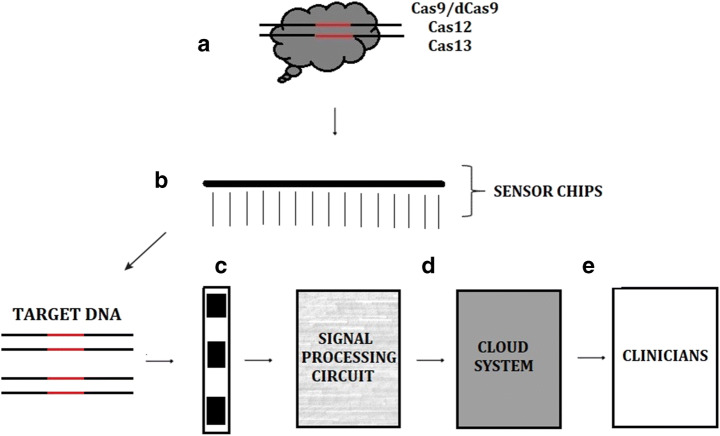
In this section, we focused our aim on designing biosensor that utilize CRISPR/Cas systems as biological recognition element, fabrication of biosensor using nanotechnology, wireless transmission of data through the cloud system or database using IoT and development of biosensor enable with AI that can be used for analysis such as prediction, support clinical decision, real-time and POC diagnosis. The overall architecture of the proposed Biosensor is shown in Figure 5.
Fig. 5.
Proposed Biosensing system
Measurement of different level of extracellular and intracellular metabolite, chemical, DNA hybridization and DNA or RNA Cleavage of different Cas effectors can be detected using growing toolbox of developed biosensors. The proposed futuristic biosensor is presented in the following steps:
Type of biosensor
There are different types of biosensors as mentioned in Table 3. The proposed future biosensors will utilize nanoelectronics biosensors integrated with CRISPR/Cas system to monitor the DNA or RNA guided bound Cas protein such as binding and cleavage and convert the molecular binding activities (such as readout results or signals) to digital stream of data.
Sensing technology
The sensing technology of the biosensor will be based on signals obtain from hybridization and cleavage of DNA or RNA which will produce signals. The signals can be linked to recording system. Prior to recording, sensed signals must be converted to specific format such as digit or analogue for easy interpretation. To achieve this conversion, signal processing circuit can be couple with the biosensor for signal computations, integration and transformation of tuning signals as reported by [26, 85]. Biosensors can also be linked with readout circuit amplification channels with a microcontroller to sense signals and generate data. Another alternative is coupling of biosensors with wireless devices or technology through the use of GPS, Wi-Fi and Bluetooth.
Transmission and storage
A) CRISPR-based biosensor will be designed using either CRISPR Cas9, deactivated or mutated Cas9 (dCas9), Cas12a (CPF1) and Cas13a (CPF2) with Single Guide RNA (SgRNA) that will match the target. B) The selected CRISPR/Cas system will be fabricated on a chip C). A DNA sample from suspected patient will be apply on the biosensor along with fabricated chip and the hybridization will produce color change or signal. D) The result in form of signal will be converted to digit using Signal Processing Circuit and the output will be transfer through the network or stored in cloud computing system and will be program to be public, private or hybrid E) Doctors and clinicians can be able to access result for support decision making.
The recorded data stored in cloud system or biomedical database and can be access by physicians for proper decision making and scientist for data analysis and research purposes. Direct electrical integration of bio robot (CRISPR/Cas system) with nanoscale transistor capable of sensing the interaction between the electrochemically charge molecules of CRISPR and DNA will be a powerful and effective tool for medical diagnosis and e-healthcare. Due to patient privacy, data obtained from biomedical sensors must be protected in the cloud computing architecture by using data encryption which can only be accessible by selected personnel such as doctors, clinicians and patients except for retrospective study.
Data analysis
Artificial intelligence and machine learning models have been integrated into medicine for data analysis based on pattern recognition, modeling and prediction. The data obtained from biosensor can be in the form of graphics (i.e. images) or numeric. For image dataset, CNN models such as ResNet, GoogleNet, VGGNet and AlexNet can be used for classification. For prediction, machine learning algorithms such as Linear and Logistic regression, Clustering algorithms, Naïve-Bayes, and K-Nearest Neighbors (KNN) can be employed.
Effect of environmental factors on fabricated Chip
A chip is a very small part of electronic device and play essential role in detection of analyte. The fabrication of CRISPR/Cas system on nanochips will result in nanoelectronics biosensor that can detect binding and cleavage of Cas system on target DNA or RNA. However, there are possibility of environmental concerns such as climate strategy, water related risks and hazardous chemicals. The environmental performances of the applications are based on the chips embedded inside the system [96]. The nanochips proposed in this study are the type that can be embedded inside the sensor and thus contact with environmental factors such as greenhouse gases and chemicals are very unlikely. Nevertheless, exposure to high temperature and toxic chemical can affect the functioning of the chips.
Challenges and open research issues
In this paper, a detail review of current conventional biosensors and CRISPR-based biosensors were discussed. The differences between the biosensors, application, strength and weakness were highlighted. As discussed in the paper, detection of pathogenic diseases is more sensitive, accurate, rapid robust and simple using CRISPR-based biosensor equipped with single guide RNA (SgRNA) for a particular strain of organism than using conventional biosensors that utilize antibodies, enzymes, whole cells etc. [15, 62]. Even though Kaushik et al. [52] argued that a specific antibody-based nano enabled electrochemical immunosensing system offer better performance for detection of Zika virus than using CRISPR-based biosensors but it does not say for other viral and bacterial pathogens and cancer mutations.
CRISPR-based biosensors offered clinicians a new paradigm of diagnosing cancer mutations, bacterial and viral diseases and for studying gene function but this method has its own limitation which must be solve for CRISPR-biosensing to achieve its full potential. Currently, there is high need for development of biomedical miniaturized biosensing system that can be deploy for point of care (POC) application. Must of the current biosensors take longer processing and turnaround time (Pardee et al. [79] 2–6 h, Huang et al. [44] less than 1 h, Bruch et al. [20] less than 4 h, Myhrvold et al. [74] 2 h etc.).
Current CRISPR-based biosensors detect target sample at different concentration such as Huang et al. [44] (0.82aM), Hajian et al. [38] (1.7fM), Koo et al. [55] (0.54aM and 0.63aM), Li B et al. [60] (10aM), Gootenberg et al. [37] (2aM), Gootenberg et al. [36] (2aM), Myhrvold et al. [74] (90aM and 20aM) and Bruch et al. [20] (10pM). However, there is high need for developing low cost, user friendly, sensitive, portable and selective CRISPR-based biosensors that can detect target sample at a very low concentration (i.e. pM, zM and yM). The most common challenge of implantable biosensors is power consumption. To addressed this limitation, a self-powered device must be design to avoid the drainage of power for implantable devices.
Complexity and privacy of data are the two other challenges, compare to other biomedical data types (such as temperature, heartbeat rate, blood pressure, blood flow, biomedical imaging, slide images etc.), to omics (such as genomics, proteomics, transcriptomics, metabolomics etc.) data types are very different and thus require approaches for conversion into digit or analogue for easy interpretation. Due to privacy of data associated with clinical sciences (such as result of CRISPR-based diagnosis) are less readily available and more need to be done in order to overcome the ethical, societal, legal and commercial barriers that restricted the use of these data for data analysis using machine learning and storage in cloud computing and online databases.
Development of smart system for detection of diseases at an early stage and monitoring diseases and physiochemical changes will be an ideal and efficient diagnosis approach that can fill the gap between estimation of infection levels and resulting therapeutic decision. Effort must be channel toward developing CRISPR-based biosensors that can be equipped with cloud computing and big biomedical data for data storage, analysis and support decision.
Conclusions
Majority of outbreak of diseases (i.e. epidemic) affected remote and rural communities with low access to medical diagnostic tools and equipment. The use of e-healthcare or telemedicine has shown promise as a n approach for solving crisis regarding accessibility to healthcare system. The outbreak of Zika virus in 2016 has led to the development of sensitive CRISPR-based biosensor that can be used to detect different strain of this virus at low concentration. The application of IoT, big biomedical data, cloud computing, artificial intelligence and signal data obtained from CRISPR-based biosensors or nano-biosensors will provide clinical data in the cloud computing system which can be accessible to restricted personnel for support decision or public for research purpose. To achieve an ideal, smart and futuristic biosensor, so many challenges must be addressed such as increasing the sensitivity of the biosensor, complexity and privacy of biomedical data, fabrication of biosensors for use as POC molecular diagnostic tool, power consumption, coupling biosensors with wireless technologies and developing multiplexing biosensors for detection of different pathogens.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Abdullahi Umar Ibrahim, Email: Abdullahi.umaribrahim@neu.edu.tr.
Fadi Al-Turjman, Email: fadi.alturjman@neu.edu.tr.
Zubaida Sa’id, Email: zubaidas11@gmail.com.
Mehmet Ozsoz, Email: mehmet.ozsoz@neu.edu.tr.
References
- 1.Abdelhamid HN, Wu HF (2013) Multifunctional graphene magnetic nanosheet decorated with chitosan for highly sensitive detection of pathogenic bacteria. J Mater Chem B 1(32):3950–3961. 10.1039/C3TB20413H [DOI] [PubMed]
- 2.Abudayyeh OO, Gootenberg JS. Chipping in on diagnostics. The CRISPR journal. 2019;100(102):69–71. doi: 10.1089/crispr.2019.29053.oma. [DOI] [PubMed] [Google Scholar]
- 3.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aileni RM, Pasca S, Valderrama C (2015) Cloud computing for big data from biomedical sensors monitoring, storage and analyze. In: 2015 conference grid, Cloud & High Performance Computing in science (ROLCG) (pp. 1-4). IEEE. 10.1109/2FROLCG.2015.7367419
- 6.Al-Turjman F. Intelligence and security in big 5G-oriented IoNT: an overview. Futur Gener Comput Syst. 2020;102:357–368. doi: 10.1016/j.future.2019.08.009. [DOI] [Google Scholar]
- 7.Al-Turjman F, Alturjman S. Context-sensitive access in industrial internet of things (IIoT) healthcare applications. IEEE Transact Indust Inform. 2018;14(6):2736–2744. doi: 10.1109/TII.2018.2808190. [DOI] [Google Scholar]
- 8.Al-Turjman F, Baali I (n.d.)Machine learning for wearable IoT-based applications: A survey. Transactions on Emerging Telecommunications Technologies, p e3635. 10.1002/ett.3635
- 9.Al-Turjman F, Nawaz MH, Ulusar UD. Intelligence in the internet of medical things era: a systematic review of current and future trends. Comput Commun. 2019;150:644–660. doi: 10.1016/j.comcom.2019.12.030. [DOI] [Google Scholar]
- 10.Al-Turjman F, Zahmatkesh H, Mostarda L. Quantifying uncertainty in internet of medical things and big-data services using intelligence and deep learning. IEEE Access. 2019;7:115749–115759. doi: 10.1109/ACCESS.2019.2931637. [DOI] [Google Scholar]
- 11.Al-Turjman FM (2016) Towards smart ehealth in the ultra large-scale internet of things era. In: 2016 23rd Iranian conference on biomedical engineering and 2016 1st international Iranian conference on biomedical engineering (ICBME) (pp. 102-105). IEEE. 10.1109/ICBME.2016.7890938
- 12.Baeumner AJ, Schlesinger NA, Slutzki NS, Romano J, Lee EM, Montagna RA. Biosensor for dengue virus detection: sensitive, rapid, and serotype specific. Anal Chem. 2002;74(6):1442–1448. doi: 10.1021/ac015675e. [DOI] [PubMed] [Google Scholar]
- 13.Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, et al. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348(6230):36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 15.Batista AC, Pacheco LG. Detecting pathogens with zinc-finger, TALE and CRISPR-based programmable nucleic acid binding proteins. J Microbiol Methods. 2018;152:98–104. doi: 10.1016/j.mimet.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Bellazzi R. Big data and biomedical informatics: a challenging opportunity. Yearb. Med. Inform. 2014;23(01):08–13. doi: 10.15265/IY-2014-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurevgenet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 18.Brey P. Ethical aspects of information security and privacy. In: security, privacy, and trust in modern data management. Berlin, Heidelberg: Springer; 2007. pp. 21–36. [Google Scholar]
- 19.Brownlee J (2016) Machine learning mastery with Python: understand your data, create accurate models, and work projects end-to-end. Machine Learning Mastery
- 20.Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, Dincer C, Urban GA. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater. 2019;31:1905311. doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 21.Carroll D (2016) The development and use of zinc-finger nucleases. In: genome editing (pp. 15–28). Springer, New York
- 22.Chang EY, Wu MH, Tang KF, Kao HC, Chou, CN (2017). Artificial intelligence in XPRIZE DeepQ Tricorder. In MMHealth@ MM (pp. 11-18)
- 23.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15(141):20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daaboul GG, Lopez CA, Yurt A, Goldberg BB, Connor JH, Ünlü MS. Label-free optical biosensors for virus detection and characterization. IEEE J Select Topics Quantum Electron. 2011;18(4):1422–1433. doi: 10.1109/JSTQE.2011.2180516. [DOI] [Google Scholar]
- 25.Dai Y, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, Liu CC. Exploring the trans-cleavage activity of CRISPR Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew Chem Int Ed. 2019;58:17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497(7451):619. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 27.Das A, Pradhapan P, Groenendaal W, Adiraju P, Rajan RT, Catthoor F, et al. Unsupervised heart-rate estimation in wearables with liquid states and a probabilistic readout. Neural Netw. 2018;99:134–147. doi: 10.1016/j.neunet.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Diouani MF, Helali S, Hafaid I, Hassen WM, Snoussi MA, Ghram A, Jaffrezic-Renault N, Abdelghani A. Miniaturized biosensor for avian influenza virus detection. Mater Sci Eng C. 2008;28(5–6):580–583. doi: 10.1016/j.msec.2007.10.043. [DOI] [Google Scholar]
- 29.Doudna JA, Charpenteir E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 30.Duan N, Wu S, Dai S, Miao T, Chen J, Wang Z. Simultaneous detection of pathogenic bacteria using an aptamer based biosensor and dual fluorescence resonance energy transfer from quantum dots to carbon nanoparticles. Microchim Acta. 2015;182(5–6):917–923. doi: 10.1007/s00604-014-1406-3. [DOI] [Google Scholar]
- 31.East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eid A, Mahfouz MM. Genome editing: the road of CRISPR/Cas9 from bench to clinic. Exp Mol Med. 2016;48(10):e265. doi: 10.1038/emm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan YJ, Yin YH, Da Xu L, Zeng Y, Wu F. IoT-based smart rehabilitation system. IEEE transactions on industrial informatics. 2014;10(2):1568–1577. doi: 10.1109/TII.2014.2302583. [DOI] [Google Scholar]
- 34.Farzadfard F, Lu TK. Emerging applications for DNA writers and molecular recorders. Science. 2018;361(6405):870–875. doi: 10.1126/science.aat9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 36.Gootenberg JS, Abudayyeh OO, KellnerMJ JJ, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hajian R, Balderston S, Tran T, Etienne J, Sandhu M, Wauford NA, Chung J, Nokes J, Athaiya M, Paredes J, Peytavi R, Goldsmith B, Murthy N, Conboy IM, Aran K (n.d.) Field-effect transistor. Nat. Biomed. Eng. 10.1038/s41551-019-0371-x [DOI] [PMC free article] [PubMed]
- 39.Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107(1):1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendel A, Kildebeck EJ, Fine EJ, Clark JT, Punjya N, Sebastiano V, Bao G, Porteus MH. Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Rep. 2014;7(1):293–305. doi: 10.1016/j.celrep.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodgkins A, Farne A, Perera S, Grego T, Parry-Smith DJ, Skarnes WC, Iyer V. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015;31(18):3078–3080. doi: 10.1093/bioinformatics/btv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 43.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang M, Zhou X, Wang H, Xing D. Clustered regularly interspaced short palindromic repeats/ Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal Chem. 2018;90(3):2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 45.Ishino Y, Krupovic M, Forterre P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol. 2018;200(7):e00580–e00517. doi: 10.1128/JB.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacob S, Menon VG, Al-Turjman F, Vinoj PG, Mostarda L. Artificial muscle intelligence system with deep learning for post-stroke assistance and rehabilitation. IEEE Access. 2019;7:133463–133473. doi: 10.1109/ACCESS.2019.2941491. [DOI] [Google Scholar]
- 47.Jatmiko W, Arsa DMS, Wisesa H, Jati G, Ma’Sum MA (2016) A review of big data analytics in the biomedical field. In: 2016 international workshop on big data and information security (IWBIS) (pp. 31-41). IEEE. 10.1109/IWBIS.2016.7872886
- 48.Jiang F, Doudna JA. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 49.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones VM, in’t Veld RH, Tonis T, Bults RB, Van Beijnum B, Widya I, Hermens H (2008) Biosignal and context monitoring: distributed multimedia applications of body area networks in healthcare. In: 2008 IEEE 10th Workshop on Multimedia Signal Processing (pp. 820–825). IEEE
- 51.Kanaparthi S, Supraja P, Singh SG (2019) Smart, portable, and noninvasive diagnostic biosensors for healthcare. In: advanced biosensors for health care applications. Elsevier, pp 209–226. 10.1016/B978-0-12-815743-5.00007-X
- 52.Kaushik A, Tiwari S, Jayant RD, Vashist A, Nikkhah-Moshaie R, El-Hage N, Nair M. Electrochemical biosensors for early stage Zika diagnostics. Trends Biotechnol. 2017;35(4):308–317. doi: 10.1016/j.tibtech.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine learning and data mining methods in diabetes research. Comput. Struct. Biotechnol. J. 2017;15:104–116. doi: 10.1016/j.csbj.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koo B, Kim DE, Kweon J, Jin CE, Kim SH, Kim Y, Shin Y. CRISPR/dCas9-mediated biosensor for detection of tick-borne diseases. Sensors Actuators B Chem. 2018;273:316–321. doi: 10.1016/j.snb.2018.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.KosickiM TK, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazcka O, Del Campo FJ, Munoz FX. Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron. 2007;22(7):1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 59.Lenoir WF, Lim TL, Hart T. PICKLES: the database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 2017;46(D1):D776–D780. doi: 10.1093/nar/gkx993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S, Wang J (2018) CRISPR-Cas12a-assisted nucleic acid detection. Cell discovery 4(1):20. 10.1038/s41421-018-0028-z [DOI] [PMC free article] [PubMed]
- 61.Li Y, He C, Fan X, Huang X, Cai Y, Terzo O, Mossucca L (2015) HCloud, a healthcare-oriented cloud system with improved efficiency in biomedical data processing. Cloud computing with e-science applications, p 163
- 62.Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Liu L, Liu G. CRISPR/Cas multiplexed biosensing: a challenge or an insurmountable obstacle? Trends Biotechnol. 2019;37:792–795. doi: 10.1016/j.tibtech.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 64.Liu F, Choi KS, Park TJ, Lee SY, Seo TS. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011;5(2):123–128. doi: 10.1007/s13206-011-5204-2. [DOI] [Google Scholar]
- 65.Liu X, Marrakchi M, Xu D, Dong H, Andreescu S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016;80:9–16. doi: 10.1016/j.bios.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 66.Liu X, Wu S, Xu J, Sui C, Wei J. Application of CRISPR/Cas9 in plant biology. Acta Pharm Sin B. 2017;7(3):292–302. doi: 10.1016/j.apsb.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, … Xie Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. 10.1016/j.molp.2015.04.007 [DOI] [PubMed]
- 68.Ma’Sum MA, Jatmiko W, Suhartanto H (2016) Enhanced tele ECG system using hadoop framework to deal with big data processing. In: 2016 international workshop on big data and information security (IWBIS) (pp. 121-126). IEEE. 10.1109/IWBIS.2016.7872900
- 69.Makarova KS, Zhang F, Koonin EV (2017) SnapShot: class 2 CRISPR-Cas systems. Cell 168(1):328–328. 10.1016/j.cell.2016.12.038328.e1 [DOI] [PubMed]
- 70.Mannoor MS, Zhang S, Link AJ, McAlpine MC. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proceedings of the National Academy of Sciences, 107(45), 19207-19212. Molecular plant. 2010;8(8):1274–1284. doi: 10.1073/pnas.1008768107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marling C, Xia L, Bunescu R, Schwartz F (2016) Machine learning experiments with noninvasive sensors for hypoglycemia detection. In: proceedings of IJCAI workshop on knowledge discovery in healthcare data. Morgan Kaufmann publishers Inc., San Francisco, pp 1–6
- 72.Miranda OR, Li X, Garcia-Gonzalez L, Zhu ZJ, Yan B, Bunz UH, Rotello VM. Colorimetric bacteria sensing using a supramolecular enzyme–nanoparticle biosensor. J Am Chem Soc. 2011;133(25):9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohanty S, Jagadeesh M, Srivatsa H (2013) Big data imperatives: Enterprise ‘Big Data’warehouse, ‘BI’implementations and analytics. Apress
- 74.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, et al. Field-deployable viral diagnostics using CRISPRCas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nayak M, Kotian A, Marathe S, Chakravortty D. Detection of microorganisms using biosensors—a smarter way towards detection techniques. Biosens. Bioelectron. 2009;25(4):661–667. doi: 10.1016/j.bios.2009.08.037. [DOI] [PubMed] [Google Scholar]
- 76.Nehal SA, Roy D, Devi M, Srinivas T (2019) Highly sensitive lab-on-chip with deep learning AI for detection of bacteria in water. Int. J. Inf. Technol.:1–7. 10.1007/s41870-019-00363-1
- 77.Novak S. Plant biotechnology applications of zinc finger technology. In: transgenic plants. New York: Humana Press; 2019. pp. 295–310. [DOI] [PubMed] [Google Scholar]
- 78.Paiva JS, Cardoso J, Pereira T. Supervised learning methods for pathological arterial pulse wave differentiation: a SVM and neural networks approach. Int J Med Inform. 2018;109:30–38. doi: 10.1016/j.ijmedinf.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 79.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, et al. Rapid, lowcost detection of Zika virus using programmable biomolecular components. Cell. 2016;165(5):1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 80.Park J, Kim JS, Bae S. Cas-database: web-based genome-wide guide RNA library design for gene knockout screens using CRISPR-Cas9. Bioinformatics. 2016;32(13):2017–2023. doi: 10.1093/bioinformatics/btw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quan TM, Doike T, Bui DC, Arata S, Kobayashi A, Islam MZ, Niitsu K (2019) AI-based edge-intelligent hypoglycemia prediction system using alternate learning and inference method for blood glucose level data with low-periodicity. In: 2019 IEEE international conference on artificial intelligence circuits and systems (AICAS) (pp. 201-206). IEEE. 10.1109/AICAS.2019.8771604
- 82.Radke SM, Alocilja EC. Design and fabrication of a microimpedance biosensor for bacterial detection. IEEE Sensors J. 2004;4(4):434–440. doi: 10.1109/JSEN.2004.830300. [DOI] [Google Scholar]
- 83.Rauscher B, Heigwer F, Breinig M, Winter J, Boutros M (2016) GenomeCRISPR-a database for high-throughput CRISPR/Cas9 screens. NucleicAacids Res:gkw997. 10.1093/nar/gkw997 [DOI] [PMC free article] [PubMed]
- 84.Rowley T. Science imitates life. Lab animal. 2013;42(8):271–272. doi: 10.1038/laban.351. [DOI] [PubMed] [Google Scholar]
- 85.Rubens JR, Selvaggio G, Lu TK. Synthetic mixed-signal computation in living cells. Nat. Commun. 2016;7:11658. doi: 10.1038/ncomms11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savaliya R, Shah D, Singh R, Kumar A, Shanker R, Dhawan A, Singh S. Nanotechnology in disease diagnostic techniques. Curr Drug Metab. 2015;16(8):645–661. doi: 10.2174/1389200216666150625121546. [DOI] [PubMed] [Google Scholar]
- 87.Sheth RU, Wang HH. DNA-based memory devices for recording cellular events. Nat. Rev. Genet. 2018;19(11):718–732. doi: 10.1038/s41576-018-0052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song G, Jia M, Chen K, Kong X, Khattak B, Xie C, Li A, Mao L. CRISPR/Cas9: a powerful tool for crop genome editing. The crop journal. 2016;4(2):75–82. doi: 10.1016/j.cj.2015.12.002. [DOI] [Google Scholar]
- 89.Stefano GB, Fernandez EA (2017) Biosensors: enhancing the natural ability to sense and their dependence on bioinformatics. 10.12659/MSM.905800 [DOI] [PMC free article] [PubMed]
- 90.Tarouco LMR, Bertholdo LM, Granville LZ, Arbiza LMR, Carbone F, Marotta M, De Santanna JJC (2012, June) Internet of things in healthcare: Interoperatibility and security issues. In: 2012 IEEE international conference on communications (ICC) (pp. 6121-6125). IEEE. 10.1109/ICC.2012.6364830
- 91.Toga AW, Foster I, Kesselman C, Madduri R, Chard K, Deutsch EW, et al. Big biomedical data as the key resource for discovery science. J Am Med Inform Assoc. 2015;22(6):1126–1131. doi: 10.1093/jamia/ocv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ulusar UD, Al-Turjman F, Celik G (2017) An overview of internet of things and wireless communications. In: 2017 International Conference on Computer Science and Engineering (UBMK) (pp. 506-509). IEEE. 10.1109/UBMK.2017.8093446
- 93.Uppada V, Gokara M, Rasineni GK. Diagnosis and therapy with CRISPR advanced CRISPR based tools for point of care diagnostics and early therapies. Gene. 2018;656:22–29. doi: 10.1016/j.gene.2018.02.066. [DOI] [PubMed] [Google Scholar]
- 94.Van Der Woude MW, Bäumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17(3):581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vashistha R, Dangi AK, Kumar A, Chhabra D, Shukla P. Futuristic biosensors for cardiac health care: an artificial intelligence approach. 3. biotech. 2018;8(8):358. doi: 10.1007/s13205-018-1368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Villard A, Lelah A, Brissaud D. Drawing a chip environmental profile: environmental indicators for the semiconductor industry. J Clean Prod. 2015;86:98–109. doi: 10.1016/j.jclepro.2014.08.061. [DOI] [Google Scholar]
- 97.Wah TY, Gopal Raj R, Iqbal U (2018) Automated diagnosis of coronary artery disease: a review and workflow Cardiology research and practice, vol 2018, pp 2018–2019. 10.1155/2018/2016282 [DOI] [PMC free article] [PubMed]
- 98.Ye L, Wang C, Hong L, Sun N, Chen D, Chen S, Han F. Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell discovery. 2018;4(1):46. doi: 10.1038/s41421-018-0049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ymeti A, Greve J, Paul V, Lambeck, Wink T, van Hövell SWFM, TAM B, Robert R, Wijn RG, Heideman, Subramaniam V, Kanger JS. fast, ultrasensitive virus detection using a young interferometer sensor. Nano Lett. 2007;7(2):394–397. doi: 10.1021/nl062595n. [DOI] [PubMed] [Google Scholar]
- 100.Yuehong YIN, Zeng Y, Chen X, Fan Y. The internet of things in healthcare: an overview. J Ind Inf Integr. 2016;1:3–13. doi: 10.1016/j.jii.2016.03.004. [DOI] [Google Scholar]
- 101.Zhang GJ, Zhang L, Huang MJ, Luo ZHH, Tay GKI, Lim EJA, Kang TG, Chen Y. Silicon nanowire biosensor for highly sensitive and rapid detection of dengue virus. Sensors Actuators B Chem. 2010;146(1):138–144. doi: 10.1016/j.snb.2010.02.021. [DOI] [Google Scholar]
- 102.Zhang Y, Qian L, Wei W, Wang Y, Wang B, Lin P, et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth Biol. 2016;6(2):211–216. doi: 10.1021/acssynbio.6b00215. [DOI] [PubMed] [Google Scholar]
- 103.Zhou J, Deng K, Cheng Y, Zhong Z, Tian L, Tang X, Tang A, Zheng X, Zhang T, Qi Y, Zhang Y. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front Plant Sci. 2017;8:1598. doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]