Figure 1.
Validation of GLOE-Seq with Budding Yeast Genomic DNA
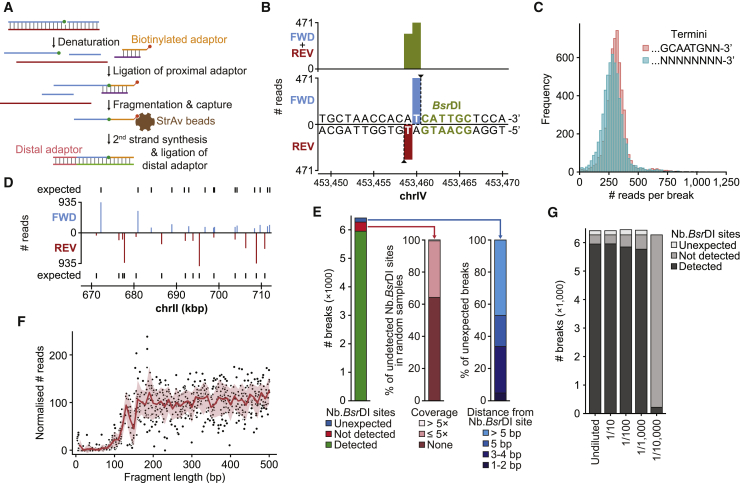
(A) GLOE-Seq workflow. Green circle, ligatable 3′-OH terminus; red circle, biotin.
(B) 3′ ends of a DSB at a BsrDI site in genome browser view (FWD, forward or Watson strand; REV, reverse or Crick strand).
(C) Histogram plot showing the distribution of read counts for the asymmetric termini generated by BsrDI.
(D) Strand-specific detection of SSBs generated by Nb.BsrDI treatment.
(E) Genome-wide detection of predicted Nb.BsrDI sites (left). Most undetected sites are absent or poorly covered in a randomly fragmented sample (center). Many of the unexpected breaks reside in the immediate neighborhood of predicted sites (right).
(F) Closely spaced SSBs are poorly detected. Nb.BsrDI signals are plotted against the calculated distance to the (upstream) neighboring site on the same strand (red, mean; pink, SE).
(G) Sensitivity of GLOE-Seq. Nb.BsrDI-treated DNA was diluted with untreated DNA at the indicated ratios. The undiluted sample corresponds to (E).