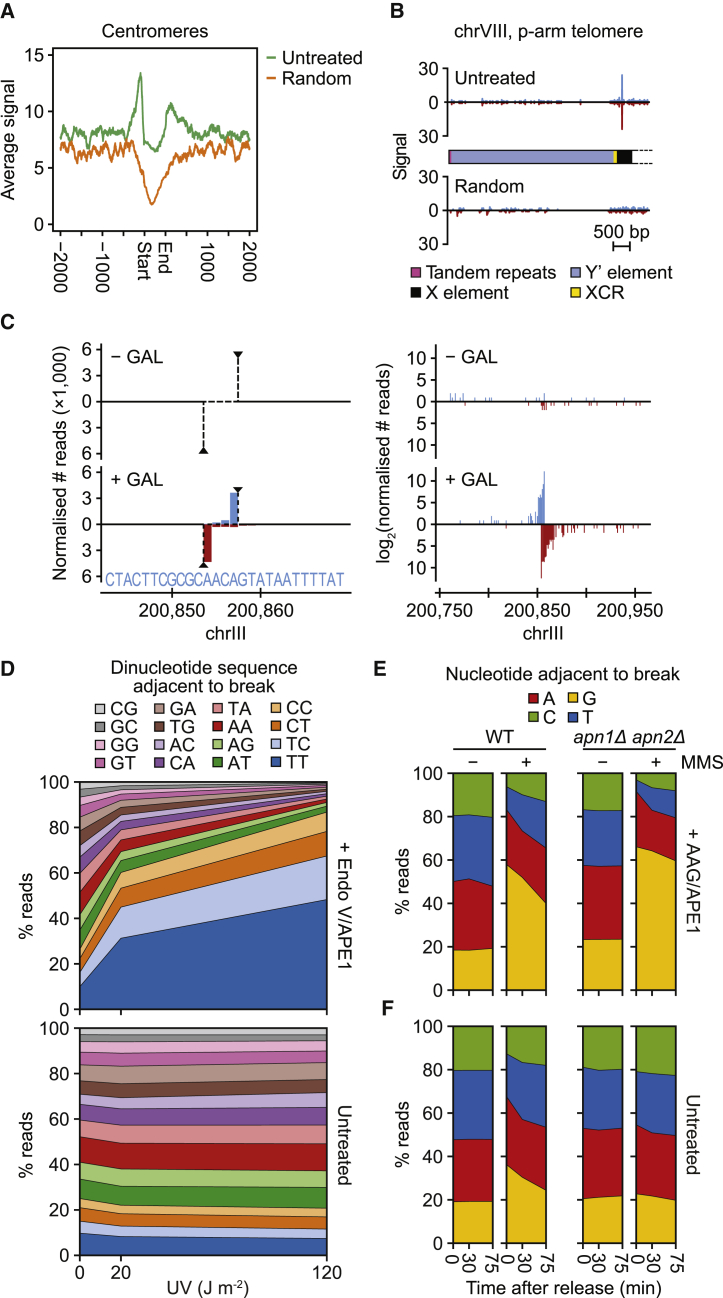
Figure 3.
GLOE-Seq Analysis of DNA Lesions and Repair Intermediates in Budding Yeast
(A) Breaks are enriched around yeast centromeres. GLOE-Seq signals of untreated and randomly fragmented DNA were averaged across all 16 centromere regions and plotted.
(B) Breaks are enriched at yeast chromosome ends. Strand-specific GLOE-Seq signals of untreated and randomly fragmented DNA are shown at a representative subtelomeric region. See Figure S4 for an image of all chromosome ends.
(C) GLOE-Seq detects the 3′ ends of a DSB, generated in vivo by galactose (GAL)-mediated induction of the HO endonuclease in yeast. Both panels show normalized numbers of reads around the HO cleavage site in a genome browser view. Left panel: linear scale at high magnification; right panel: logarithmic (log2) scale at lower magnification.
(D) GLOE-Seq detects UV irradiation-induced pyrimidine dimers in yeast. Exponentially growing yeast cultures were exposed to the indicated doses of UV radiation, and lesions were converted to strand breaks by pre-treatment of isolated genomic DNA with T4 endonuclease V and APE1 where indicated. Plots show relative frequencies of dinucleotide sequences adjacent to the detected strand breaks.
(E) GLOE-Seq detects alkylation-induced base damage in yeast. G1-arrested WT and apn1Δ apn2Δ cells were exposed to 0.02% MMS for 30 min and released into S phase in the absence of MMS. Genomic DNA was isolated from samples collected at the indicated time points, and base lesions were converted to strand breaks by pre-treatment with AAG and APE1. Plots show relative nucleotide frequencies over time during the recovery period.
(F) GLOE-Seq detects BER intermediates in yeast. Strand breaks were detected in the same samples of genomic DNA as in (E) by GLOE-Seq without AAG/APE1 pre-treatment, and relative nucleotide frequencies were plotted as in (E).