Figure 6.
GLOE-Seq Analysis of DNA Replication Patterns in Human Cells
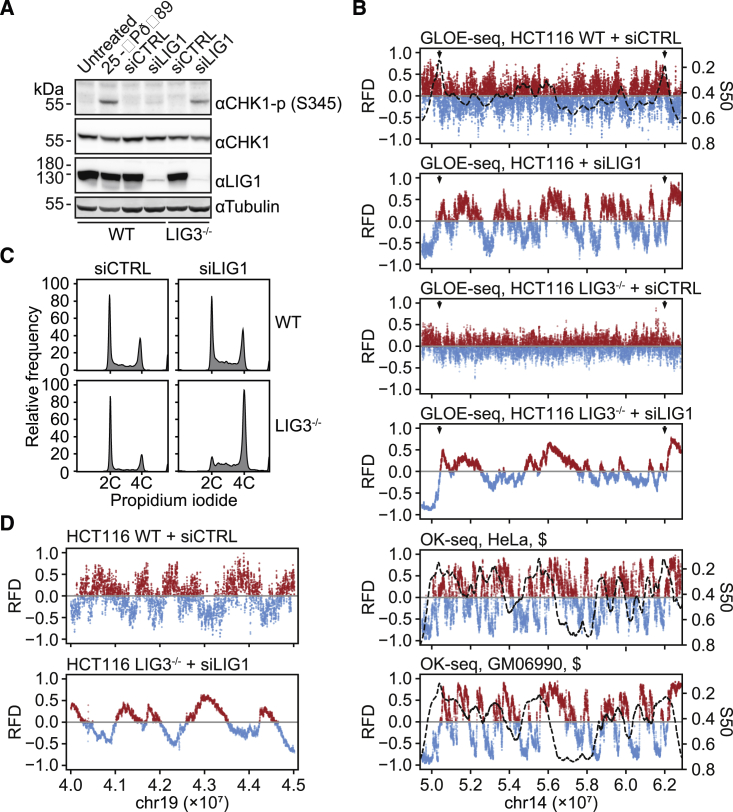
(A) Western blot images showing CHK1 phosphorylated at Ser 345, total CHK1, ligase 1, and tubulin (loading control) in whole-cell extracts prepared from HCT116 WT and LIG3−/−:mL3 cells treated with an unspecific control (siCTRL) or a ligase 1-specific siRNA (siLIG1). Treatment with 25 J/m−2 UV irradiation served as a control for checkpoint activation.
(B) RFD plots of GLOE-Seq data from HCT116 cells reproduce replication patterns under conditions of DNA ligase inactivation (HCT116 WT and LIG3−/−:mL3 cells treated with siCTRL or siLIG1, as in A). Data represent averages of two independent experiments. Replication timing (S50, dashed lines) was modeled in HCT116 WT using genome-wide DNase I hypersensitivity data from the ENCODE project (Data S1). Arrowheads indicate early and strong replication initiation zones. §, OK-Seq-derived RFD data and S50 profiles (dashed lines) from Petryk et al. (2016), shown for comparison.
(C) Flow cytometry analysis of nuclei prepared for GLOE-Seq from the same set of cells as in (A).
(D) Opposite strand biases in the break patterns of HCT116 ligase-competent (WT) versus ligase-deficient (LIG3−/−:mL3 + siLIG1) cells, illustrated by RFD plots.