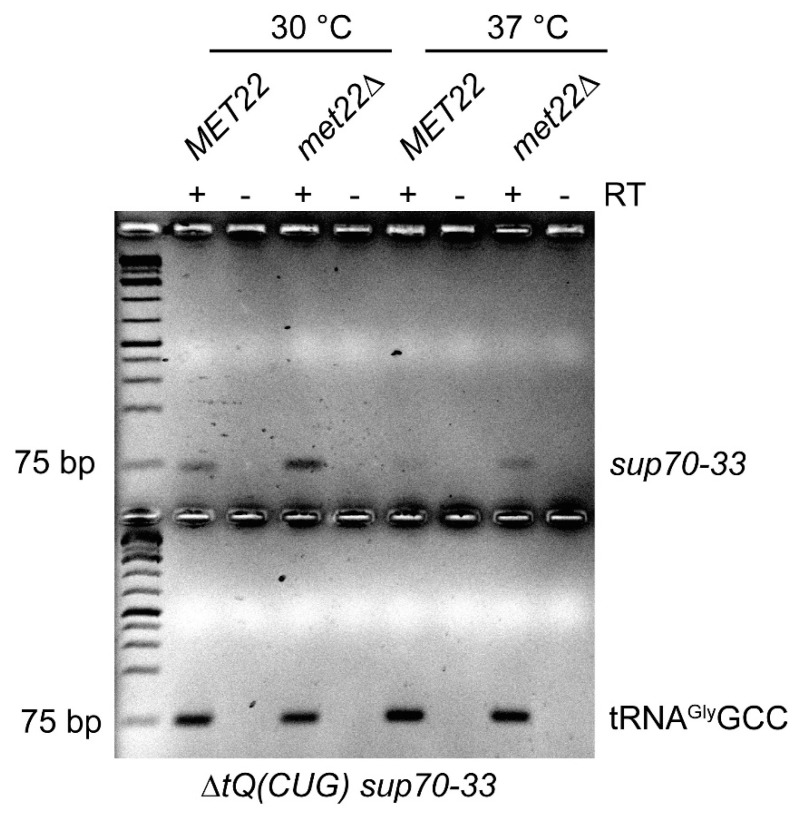
Figure 3.
Impact of met22 mutation on sup70-33 abundance. Total RNA was isolated from sup70-33 mutants with or without the met22 mutation, grown at 30 °C or shifted to 37 °C for 5 h as indicated. Identical amounts of RNA were reverse-transcribed (RT+) to cDNA using oligonucleotides specific for sup70-33 and tRNAGlyGCC, respectively. For control purposes, identical reactions were carried out omitting the reverse transcriptase (RT-). Low-cycle-number PCR was used to amplify cDNAs from sup70-33 and tRNAGlyGCC and reactions were separated on a 2% agarose gel. The 75 bp band of the DNA marker used is indicated.