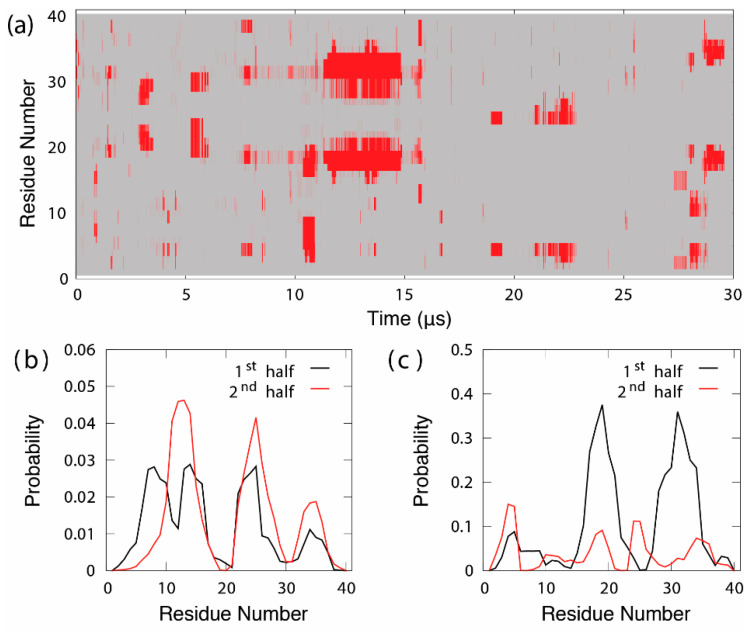
Figure 1.
(a). Evolution of the per-residue β-sheet structure during a 30-μs Anton MD simulation of intrinsically disordered Aβ40 in explicit solvent at 300 K. Residues assigned to be in the β-sheet conformation are colored in red. (b) Average residue helix and (c) β-sheet probability profiles derived from the first and second halves of the trajectory. Note that both pairs of profiles differ significantly, reflecting a lack of convergence in the simulated disordered ensemble. The original MD trajectory was generously provided by D. E. Shaw Research [92].