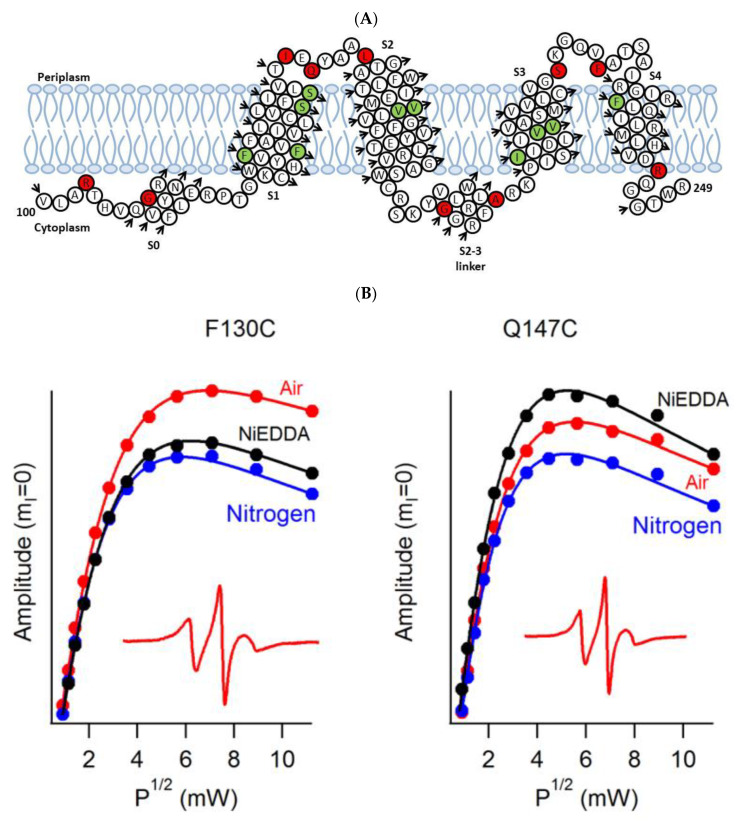
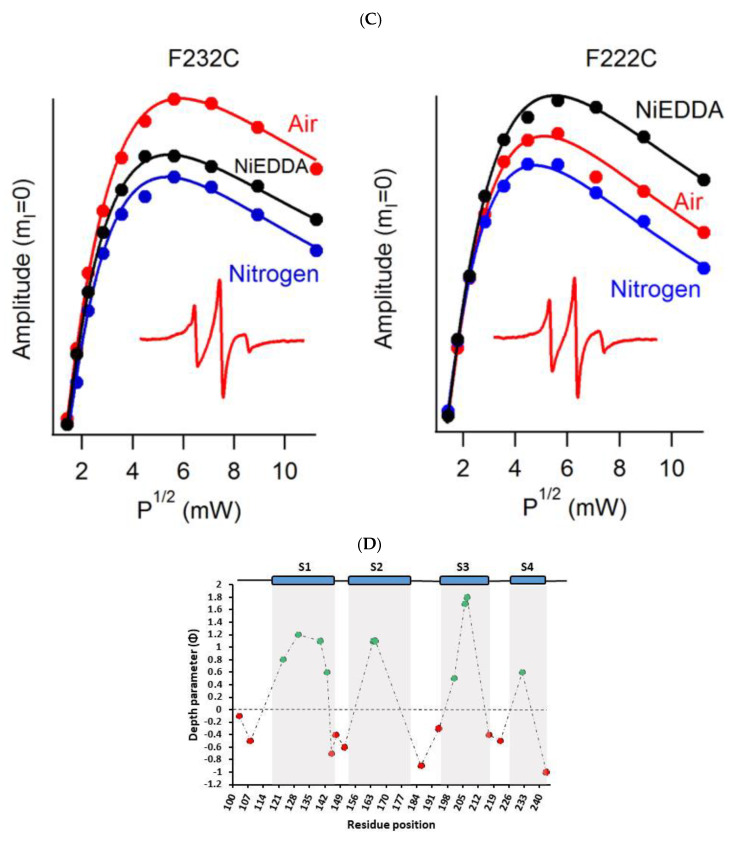
Figure 5.
(A) The proposed topology of the Q1VSD sequence in lipid bilayers. The black arrows show the order of the amino acid residues in the protein sequence. The green and red circles represent spin-label sites buried inside and outside of the membrane bilayers respectively. (B) and (C) EPR power saturation curves from Q1VSD in 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG) lipid-bilayered vesicles at 295 K. Mutation F130C is on helix S1 and is a part of the transmembrane domain, while the Q147C site is at the linker between helix S1 and helix S2 at a site outside the lipid bilayer. Mutation F232C is on helix S4 and is a part of the transmembrane domain, while the F222C site is at the linker between helix S3 and helix S4 at a site outside the lipid bilayer. The inset spectra are the corresponding CW-EPR spectra for these sites. (D) Membrane depth parameter (ϕ) as a function of Q1VSD residue position in POPC/POPG lipid-bilayered vesicles at 295 K. (Adapted from [84] with permission).