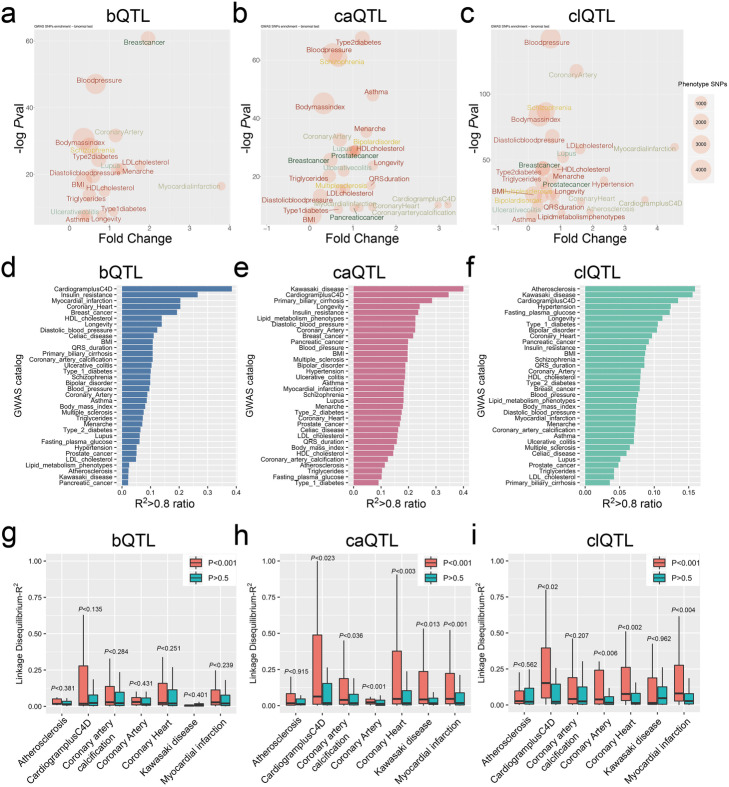
Fig. 2.
QTLs are highly associated with GWAS CAD loci. a bQTL, b caQTL, and c clQTL plots show the lead SNPs of GWAS catalog enrichment in the ± 1-kb windows of the QTLs, including P values, fold changes, and number sizes. The colors of the text indicate the different categories of diseases. d bQTL, e caQTL, and f clQTL bar graphs show the ranked GWAS terms for associated lead SNP-QTL pairs with linkage disequilibrium (LD) (R2 > 0.8) in the GWAS catalog, compared to the total number of pairs. g bQTL, h caQTL, and i clQTL box plots show the total LD R2 distributions of QTL to lead SNP pairs, in GWAS CAD-associated diseases