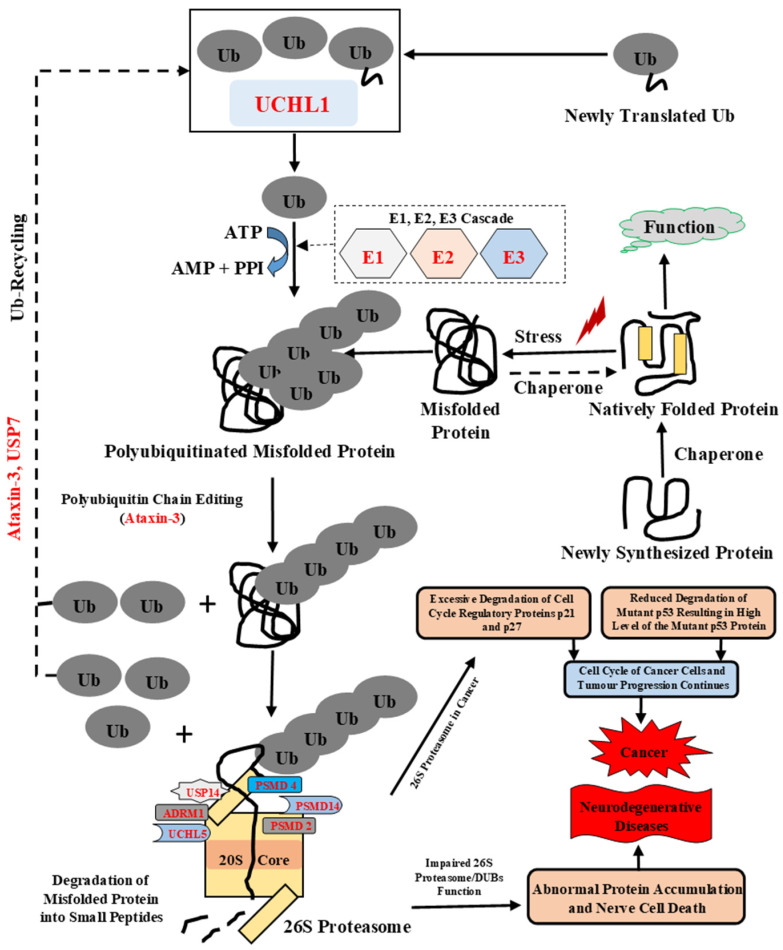
Figure 1.
Schematic representation of the ubiquitin proteasomal system. Ubiquitination is an ATP-dependent process performed by three enzymes: E1 (Ub-activating) enzyme, E2 (Ub-conjugating) enzyme, and E3 (Ub-ligase) enzyme. The DUBs, such as ataxin-3, modify the polyubiquitinated chain, to confirm accurate recognition of the misfolded proteins by the 26S proteasome. This covalent modification of misfolded protein targets them to multicatalytic protease complex, the 26S proteasome. Ubiquitination is reversed by DUBs and disassembles polyubiquitin chains. DUBs such as USP7, UCHL1, and ataxin-3 also control and maintain free Ub molecules in the cell. UCHL1 modifies newly translated protein and maintains a pool of mono-Ub. The polyubiquitinated misfolded protein can bind either to the Ub receptor of the 19S regulatory complex or to an adaptor protein that consists of both poly-Ub binding and proteasome binding domain [27]. Once misfolded protein binds to proteasome, the unfolding of the misfolded protein occurs by ATPases followed by removal of the poly-Ub chain by proteasome-associated DUBs and further translocation and degradation of unfolded protein in central proteolytic chamber occurs. Excessive degradation of cell-cycle-regulatory proteins such as p21 and p27 and reduced degradation of mutant p53 leads to a continuous cell cycle of cancer cells and tumor progression leads to the development of cancer [28]. Additionally, impairment in function of 26S proteasome, ubiquitinating enzymes, and DUBs can lead to nerve cell death and the progression of neurodegenerative diseases. Ub: Ubiquitin, E1: Ub-activating enzyme, E2: Ub-conjugating enzyme, E3: Ub-ligase enzyme.